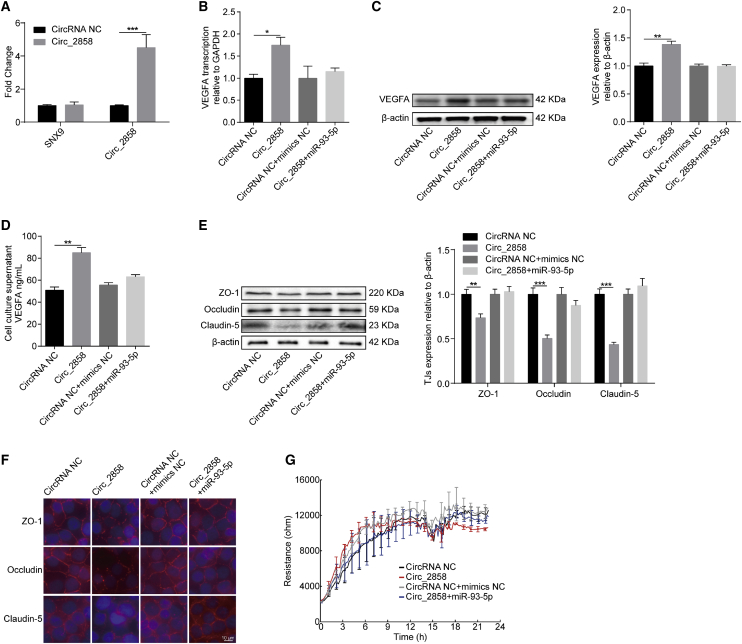
Figure 6.
Involvement of the circ_2858/miR-93-5p Axis in VEGFA-Mediated BBB Damage
(A) qPCR analyzing the transcription of circ_2858, and its parental gene SNX9, in hBMECs with circ_2858 overexpression. Data are expressed as mean ± SD from three independent experiments. (B) The expression of VEGFA in circ_2858-overexpressing hBMECs co-transfected or not with miR-93-5p determined by qPCR analysis. (C) The expression of VEGFA in circ_2858-overexpressing hBMECs co-transfected or not with miR-93-5p, as determined by western blot analysis. β-actin was used as loading control. (D) ELISA determining the secreted VEGFA levels in circ_2858-overexpressing hBMECs co-transfected or not with miR-93-5p. Data were collected and calculated as mean ± SD from three independently replicated wells. (E) Expression of TJ proteins in circ_2858-overexpressing hBMECs co-transfected or not with miR-93-5p determined by western blot. β-actin was used as loading control. (F) Distribution of TJ proteins in circ_2858-overexpressing hBMECs co-transfected or not with miR-93-5p, as determined by immunofluorescence analysis. Scale bar, 10 μm. (G) ECIS system monitoring TEER changes of the circ_2858-overexpressing hBMECs co-transfected or not with miR-93-5p. Data were collected and are presented as mean ± SD from three independently replicated wells at each time point.