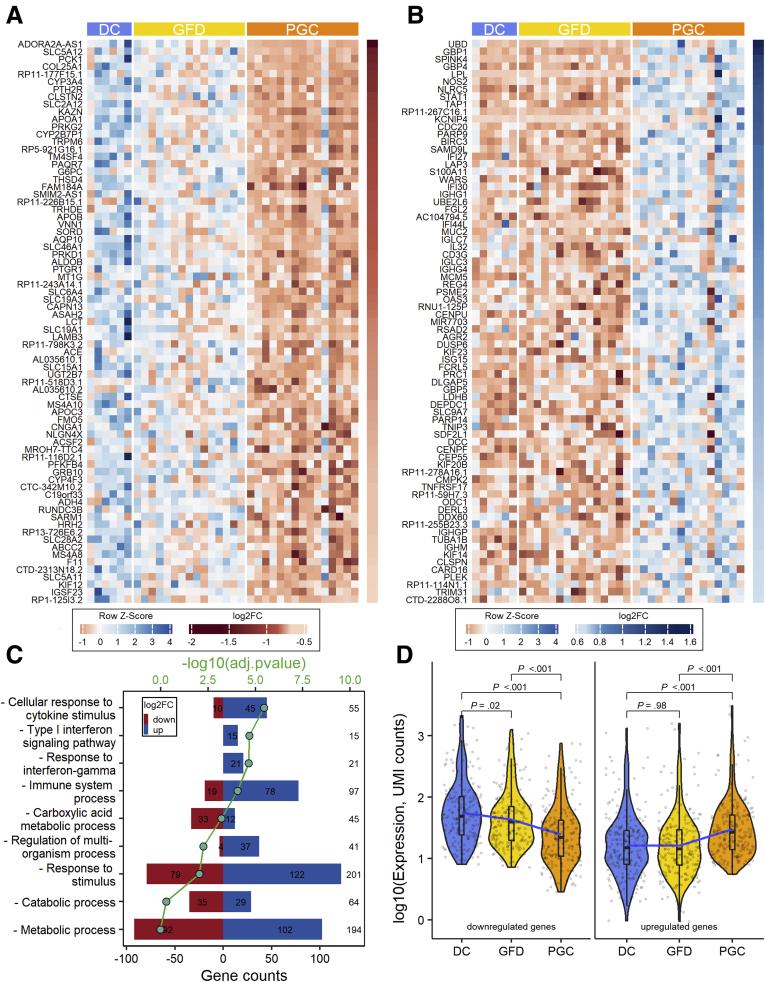
Figure 3.
Differential intestinal mucosal gene transcriptome after 10 weeks of the gluten challenge in celiac disease patients. (A) Heatmap of the top 75 genes that are downregulated specifically after the gluten challenge. (B) Heatmap of the top 75 genes that are upregulated after the gluten challenge. (C) Selected Gene Ontology terms enriched in identified set of genes (total 417 genes) specific for gluten-induced mucosal damage. (D) Violin plots showing the log-transformed expression of differentially expressed genes detected in PGC vs GFD comparisons (195 downregulated in the left panel and 222 upregulated genes in the right panel) in each group. Three lines (from the bottom to the top) in each violin plot show the location of the lower quartile, the median, and the upper quartile, respectively. The blue line is the fitted regression model, demonstrating the nonlinear character of mean of the log-transformed expression changes. Student’s t test was used for group mean comparisons.