FIG 1.
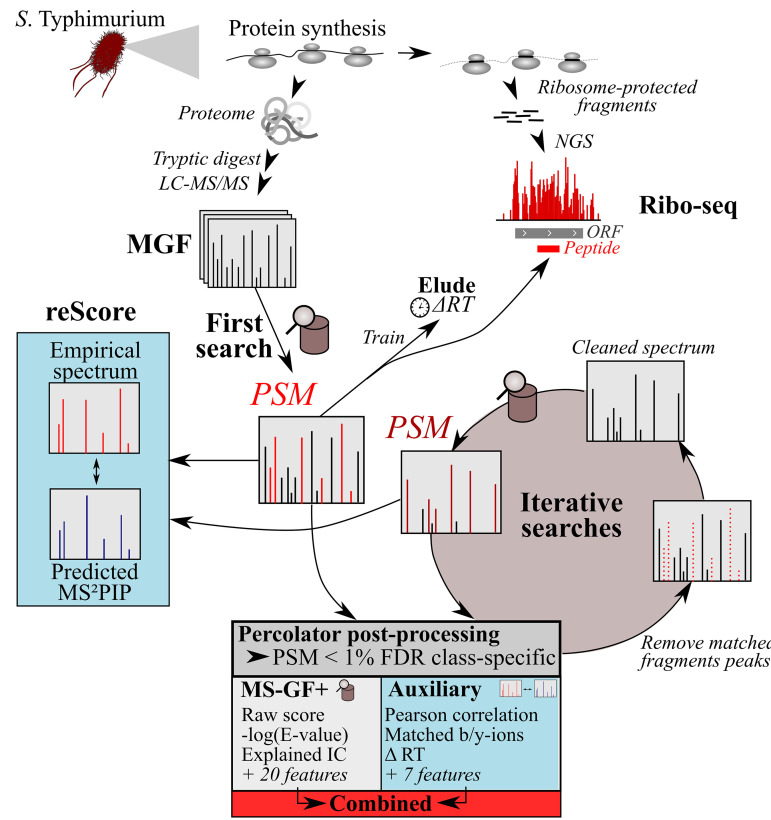
Proteomics pipeline using Percolator (15) postprocessing of 34 features (Table S1). S. Typhimurium protein expression was studied by ribosome profiling (ribo-seq) data (12) and proteomic shotgun analysis. Spectra were searched by MS-GF+ against an in silico-digested tryptic peptide database (see the text). The retention times (RT) of the top-scoring 1,000 nonredundant peptides (highest MS-GF+ score) were used to train an RT model with ELUDE (28) and calculate the deviation of empirical and predicted RT (ΔRT). Besides ΔRT, an additional 10 PSM quality features were measured, constituting the auxiliary feature set. The MS-GF+, auxiliary, or combined feature set was used by Percolator (15) for re-scoring of PSMs. Q values were re-estimated in a class-specific manner for annotated and novel peptides. Identified fragment ions were removed from spectra with a significant PSM (Q value < 0.01; combined feature set) and searched iteratively to identify cofragmented peptides. Per search, identical search and postprocessing steps were repeated as for the first search, except that the trained RT model was used from the first search and a wider precursor mass tolerance was applied (as described by Shteynberg et al. [26]).