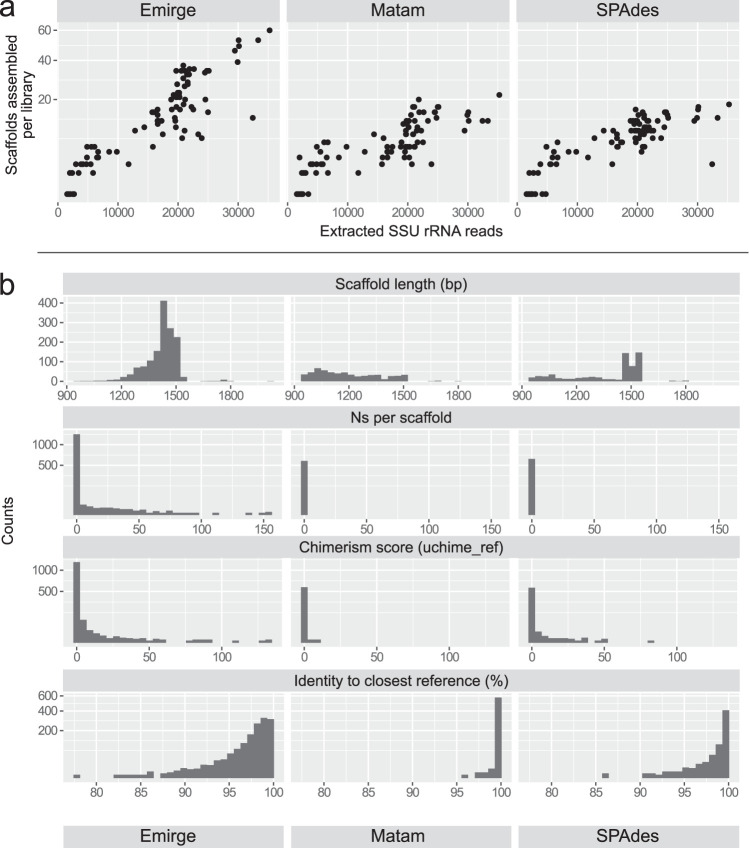
FIG 5.
Comparison of scaffolds assembled by SPAdes, Emirge, and Matam using reads extracted by SortMeRNA (E-value of 10−5, filtered reference database) from 85 Tara Oceans metagenomic libraries. (a) Plot of scaffolds assembled per library versus number of extracted SSU rRNA reads. (b) Histograms of per-scaffold metrics for each assembly tool: scaffold length (bp), undetermined bases (Ns) per scaffold, uchime_ref chimerism score, and identity to closest reference sequence.