Figure 2. The Binding Stability of CBX2 Is Independent of PRC1 and PRC2.
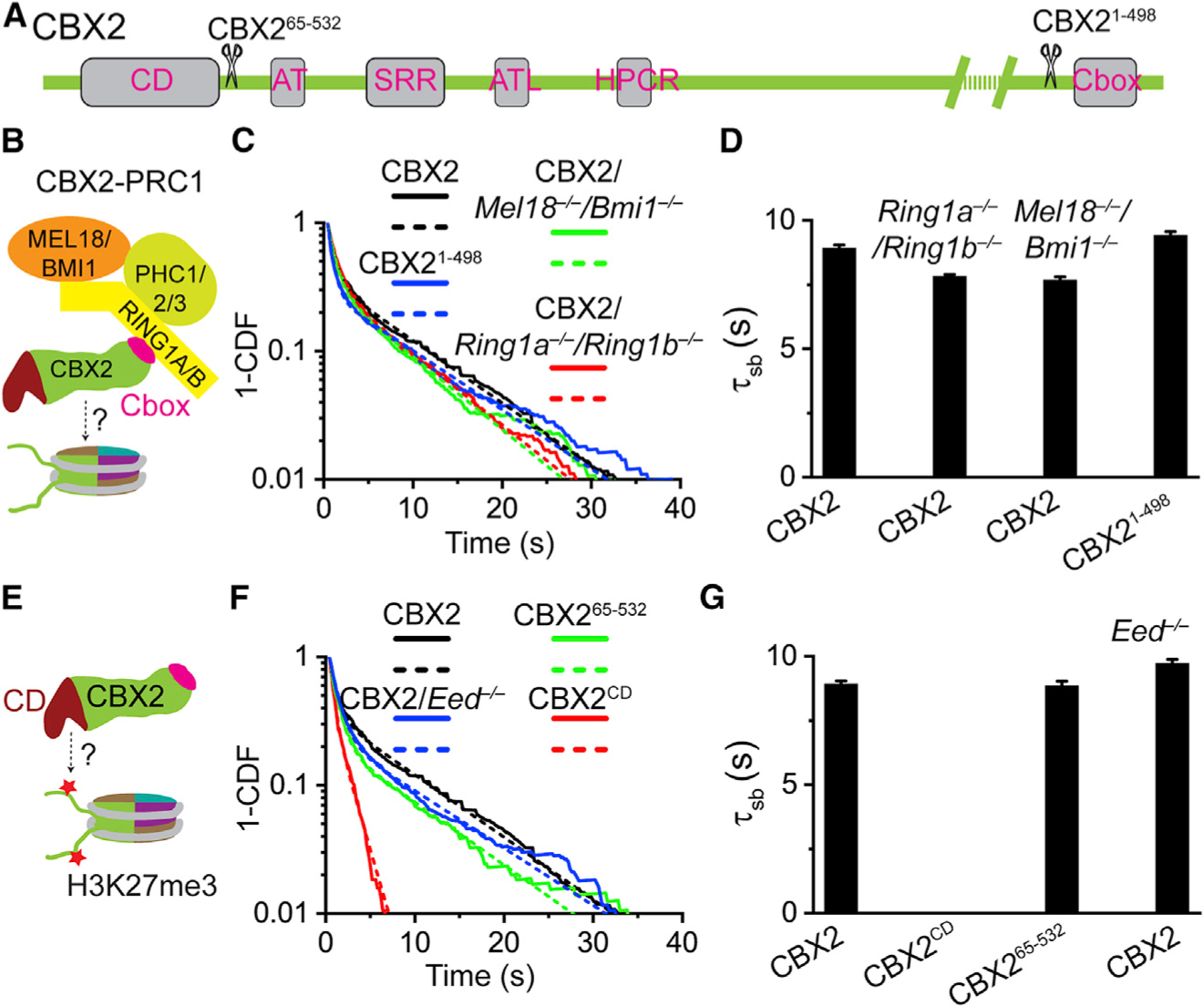
(A) Schematic representation of CBX2 and its variants. CD denotes chromodomain; AT, AT-hook; SRR, serine-rich region; ATL, AT-hook-like; HPCR, highly positively charged region; and Cbox, Chromobox.
(B) Sketch of the CBX2-PRC1 complex. The Cbox motif of CBX2 interacts with RING1B.
(C and F) Survival probability distribution of the dwell times. The numbers of cells and trajectories used are listed in Table S1.
(E) The hypothesis tests whether H3K27me3 affects the binding stability of CBX2 through interaction with the CD motif of CBX2.
(D and G) Specific residence times (τsb) quantified from (C) and (F). Non-specific residence times (τtb) are shown in Figure S2. Error bars represent standard error for the derived parameter.
