Figure 7. LLPS Alters the Target-Search Pathway.
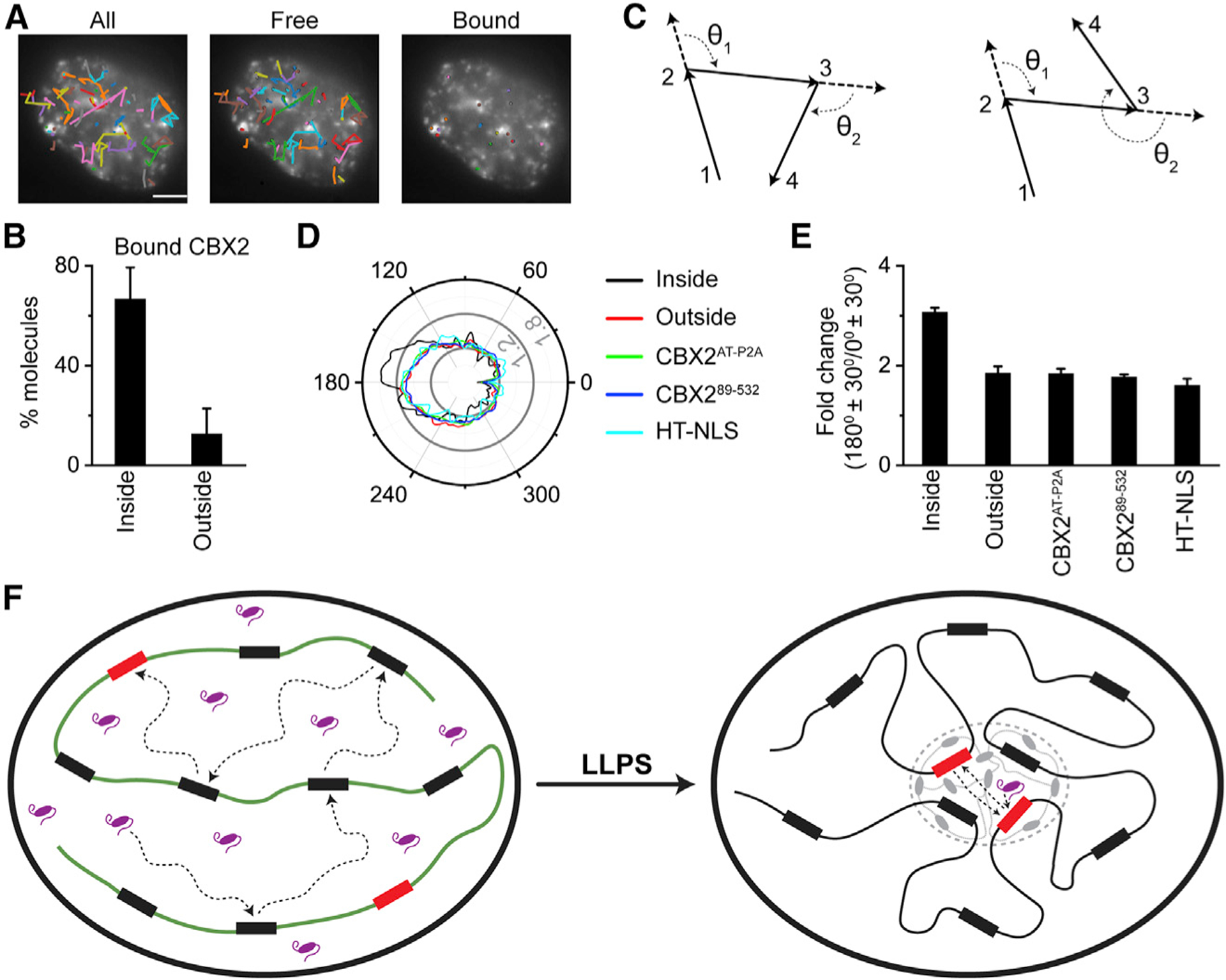
(A) Representative overlay images of SMT trajectories on epifluorescence images of CBX2. The images are represented as total trajectories (left), trajectories not bound to chromatin (middle), and trajectories bound to chromatin (right). The black circles indicate the start position of trajectories. The colors of trajectories are randomly assigned for each image. Scale bar, 5.0 μm.
(B) Percentile of chromatin-bound CBX2 molecules that are inside and outside of condensates. Error bars represent SD.
(C) Examples of the angular distribution between consecutive steps of single-molecule tracking traces.
(D) Representative angular distribution for diffusive CBX2 inside and outside of condensates as well as for CBX2AT-P2A, CBX289–532, and HT-NLS in whole cells. The major ticks of radial scale are 0.6%, 1.2%, and 1.8%.
(E) Quantification of the relative probability of moving backward compared with moving forward ([180° ± 30°]/[0° ± 30°]) for diffusive CBX2 inside and outside of condensates as well as for CBX2AT-P2A, CBX289–532, and HT-NLS in whole cells. Error bars represent SD.
(F) A proposed mechanism underpinning that CBX2 undergoes LLPS to form condensates, which then speeds up the target-search kinetics of CBX2, thereby enhancing its genomic occupancy. Our data indicate that phase-separated condensates shorten the target-search process through reducing the 3D free diffusion time and the number of non-specific trials.
