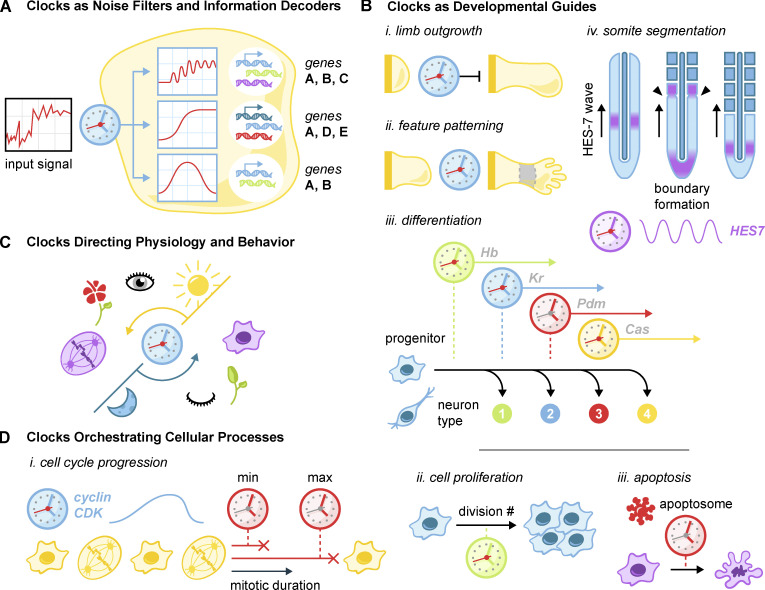
Figure 5.
Uses of biological clocks. (A) Clocks are used as noise filters and information decoders in intercellular signaling networks. Kinetic filtering and timed negative feedback programs convert noisy input signals into acute, sustained, or oscillatory pathway activation. These dynamic responses elicit different gene expression patterns. (B) Varied clock usage in development. (i) Clocks determine the outgrowth size of developing limbs. (ii) Clocks act in concert with morphogen gradients to specify digits as well as particular developmental zones. (iii) Temporal sequential expression and decay of transcription factors in neural progenitor cells lead to the specification of multiple sets of differentiated neurons. (iv) Oscillations of HES genes traveling along the developing paraxial mesoderm intersect with a wavefront to generate segmented somites. (C) Circadian oscillations direct a wide variety of behaviors such as sleep/wake, cell division, and flower blooming. (D) Clocks manage many basic cellular processes. (i) The cell cycle is driven by the cyclin/CDK oscillator: a minimum- and maximum-duration clock set the appropriate duration of mitosis. (ii) Clocks limit programmed proliferative bursts such as that which occurs in lymphocytes following infection. (iii) The procaspase-9 clock limits the duration of apoptosome activity in programmed cell death.