Figure 3.
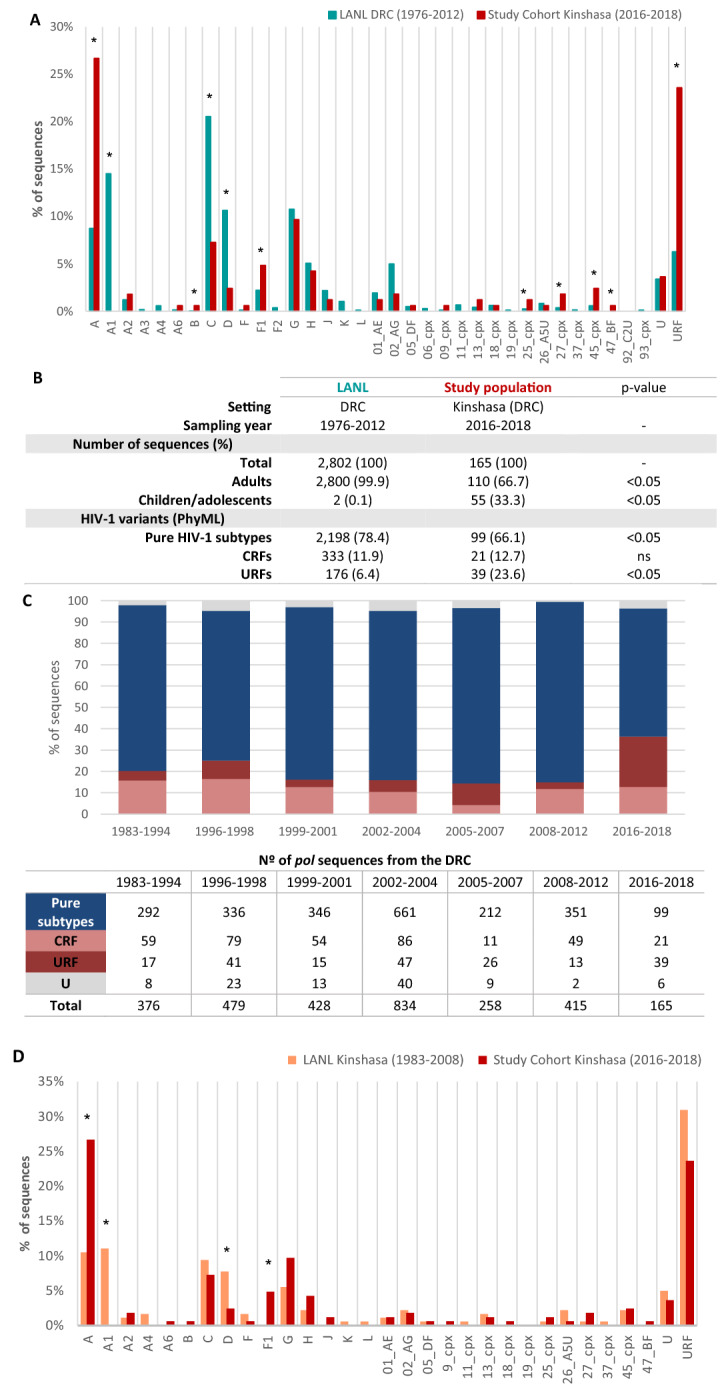
Comparison of HIV-1 variants in the study cohort (2016–2018) versus LANL sequences from the DRC (1976–2012) and Kinshasa (1983–2008). (A) Comparison of HIV-1 variants in the study cohort from Kinshasa (2016–2018) versus DRC sequences available in LANL (1976–2012). (B) Comparison of data between study cohort and LANL. (C) Evolution of pure subtypes and recombinants forms in the DRC by periods of time during 1983–2018. Panel C excludes pol sequences from the DRC sampled during 1977–1982, 1986, 1995, 2009, 2010, 2013–2015, due to their absence in LANL database. We clustered all sequences sampled during 1983–1994 as an “early period” to avoid biases due to the non-representative number of pol sequences each year within that period. Panels B and C exclude 214 LANL sequences from the DRC with unknown sampling time collected before 2012. (D) Comparison of HIV-1 variants distribution circulating in Kinshasa during 1983–2008 versus 2016–2018. For analysis we used the 181 pol sequences with available GenBank accession number from samples collected in that city during 1983–2008 versus the new 165 pol sequences collected during 2016–2018 reported in this study. Cpx, complex; CRF, circulating recombinant form; URF, unique recombinant form. *p value < 0.05.
