Figure 2.
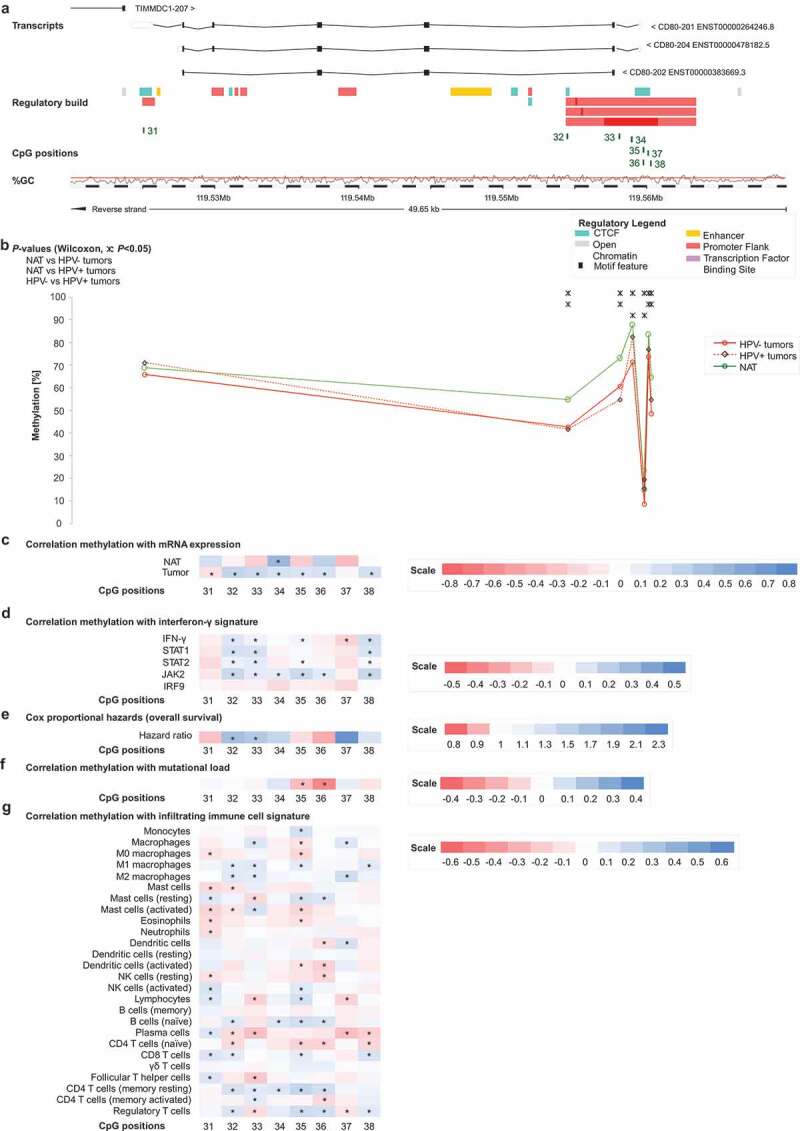
Genomic organization of CD80 (a); CD80 methylation in normal adjacent and tumour tissues (b); correlations with mRNA expression (c), interferon-γ signature (d), overall survival (e), mutational load (f), and tumour infiltrating immune cells (g). a) Shown are CG-density and eight target sites of HumanMethylation450 BeadChip beads. CpGs 31–38 (31–32: gene body, 33–34: central promoter, 35–38: promoter flank). b) Shown are methylation levels within the eight targeted CpG loci within CD80 in normal adjacent tissue (NAT), HPV+ and HPV– tumour tissue. c) Shown are Spearman’s ρ of CD80 methylation and CD80 mRNA expression and d) with interferon-γ signature. e) Cox proportional hazards with regard to CD80 methylation. f) Correlations between CD80 methylation and mutational load. g) Correlations between CD80 methylation and signatures of tumour infiltrating leukocytes. Statistically significant features are marked with asterisks. P-values refer to Wilcoxon-Mann-Whitney U test for comparisons, to Spearman’s ρ for correlations, and Wald test for Cox proportional hazard analysis
