Figure 2.
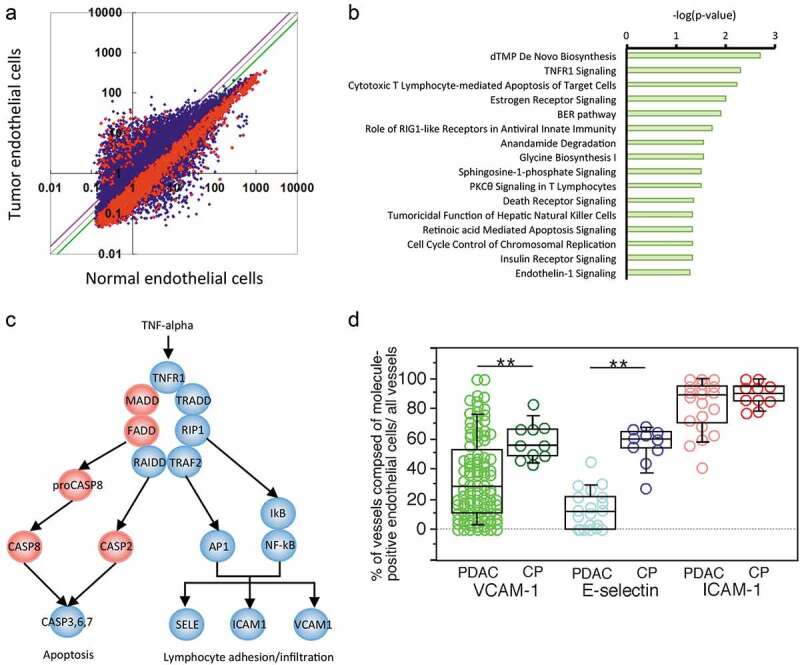
Gene expression and pathway profiles of TECs. (a) Scatter plot analysis of microarray datasets depicting the different expression profiles of TECs and endothelial cells isolated from CP tissue. The X- and Y-axes indicate averaged signal values. In total, 24,456 probes were examined, and the red dots indicate genes with expression that was significantly different between the two groups, whereas the blue dots indicate genes with no significant difference in expression. The purple and green lines indicate borders of 1.5- and 0.67-fold changes, respectively; 326 genes exhibited upregulated expression, and 2,422 genes exhibited downregulated expression in the TECs compared to the CP endothelial cells. (b) Ingenuity pathway analysis (IPA) of the altered pathways between TECs and CP endothelial cells. The pathways are ranked from the lowest P-value (top) to the highest (bottom). The green bars indicate the – log(P-value) of each pathway, so the strength of each statistical association is represented by the length of the corresponding bars. (c) Visualization of altered TNFR1 signaling in TECs. IPA canonical pathway analysis reconstructed TNFR1 signaling. The proposed intracellular signaling is illustrated. The genes with significantly upregulated expression in TECs are colored red, and those with significantly downregulated expression in TECs are colored blue. (d) The expression status of TECs in PDAC tissue samples compared to that in CP tissues determined by immunohistochemistry. Box plots show the value distribution of the percentage of vessels composed of VCAM-1+, E-selectin+, and ICAM-1+ endothelial cells among all vessels detected by evaluating ERG1+ or vVF+ endothelial cells in PDAC [VCAM-1 (n = 104), E-selectin (n = 20), and ICAM-1 (n = 20)] and CP tissue (n = 10). The scatter blots are combined. Whisker heights extend to 1.5 times the height of the box or, if no case/row has a value in that range, to the minimum and maximum values. The horizontal lines within the box indicate the median values. Differences were analyzed by using the Mann-WhitneyU test. *, P < .05 and **, P < .01
