ABSTRACT
Ten-eleven Translocation (TET) proteins have emerged as a family of epigenetic regulators that are important during development and have been implicated in various types of cancers. TET is a highly conserved protein that has orthologues in almost all multicellular organisms. Here, we review recent literature on the novel substrate specificity of this family of DNA 5-methylcytosine demethylases on DNA 6-methyladenine and RNA 5-methylcytosine that were first identified in the invertebrate model Drosophila. We focus on the biological role of these novel epigenetic marks in the fruit fly and mammals and highlight TET proteins’ critical function during development specifically in brain development.
KEYWORDS: Drosophila, epigenetics, DNA demethylation, neuroscience, cancer biology
Introduction
Epigenetic processes are critical during development and are required to maintain a balanced state of gene expression in a tissue- and cell-specific manner. Alterations in the epigenetic landscape is one main feature of tumorigenesis and is categorized as a hallmark of cancer [1].
DNA methylation is among the most widely studied and well-understood epigenetic modifications, especially methylation of cytosine on carbon 5 (5-methylcytosine). Interestingly, methylation of adenine on carbon 6 (6-methyladenine) on DNA was previously thought to be found in prokaryotes only, but recent studies point towards its presence in a variety of eukaryotic systems [2–4]. Notably, RNA modifications have been known to be more prevalent and diverse than DNA modifications for several decades [5], but their functional roles in transcription and translation have only recently emerged with the advent of transcriptome-wide analysis using NGS techniques [6]. While the presence of 5-methylcytosine (5mC) in eukaryotic RNA molecules has been reported years ago [7,8], the discovery of 5-hydroxymethylcytosine on mRNA (5hmrC), a product of the active demethylation of 5-methylcytosine on mRNA (5mrC), as an independent epigenetic mark occurred only recently [9]. Drosophila melanogaster has served as an exemplary model system in which these two modifications can be studied. The recent discovery of the demethylating enzyme dTet in fruit flies has enriched our understanding of the role of TET in development and disease. This review will cover recent discoveries on dTet and describe the relevance of both RNA and DNA modifications by dTet in development.
DNA methylation in Drosophila
DNA methylation of 5-cytosine plays an important role in gene regulation and cellular differentiation in animals and plants. It is crucial in various processes including regulation of tissue-specific gene expression, establishment and maintenance of genomic imprinting and X chromosome inactivation, as well as transcriptional repression of retrotransposons [10]. DNA methylation occurs by conversion of cytosine bases to 5-methylcytosine with the help of DNA methyltransferase (DNMT) enzymes. Demethylation of cytosines can either be an active process catalysed by TET enzymes (Figure 1b) or passive by dilution [11]. In mammals, DNA methylation is dynamic during early development with two waves of demethylation and remethylation affecting the entire genome [12]. DNA methylation in differentiated mammalian tissues is almost exclusively found in CpG dinucleotides and 70–80% of all CpG sites are methylated. The global methylation pattern of different tissues shows similar trends; while there are some highly methylated features including satellite DNA, repetitive elements such as transposons, non-repetitive intergenic DNA, and exons of genes, most sequences are methylated according to their CpG dinucleotide frequency. In contrast to that, promoter-associated CpG islands and first exons are largely unmethylated. In fact, about 75% of all promoters are estimated to lie within unmethylated CpG islands [10,13].
Figure 1.
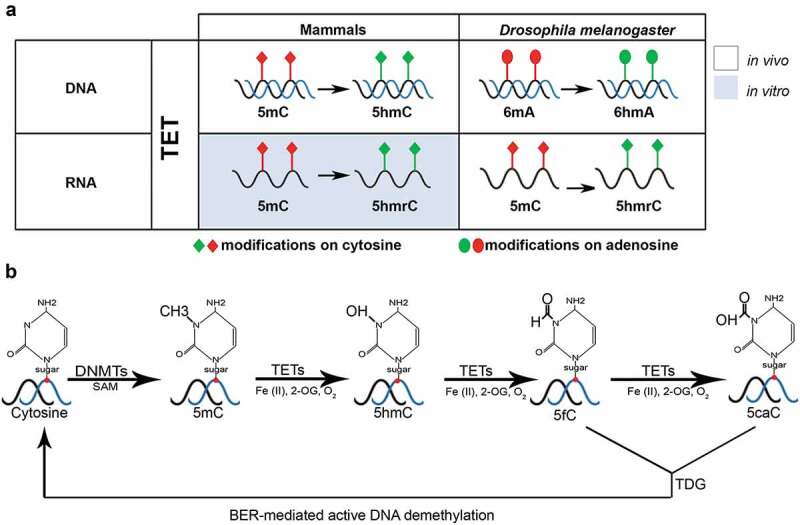
TET-driven demethylation on different substrates (RNA/DNA) in mammals and D. melanogaster. (a) TET in mammals and D. melanogaster demethylates the DNA on cytosines and adenosines, respectively. RNA demethylation is also carried out by TET in D. melanogaster; however, it has only been investigated in vitro in mammals. (b) A schematic representation of active DNA demethylation by TET, TDG (thymidine-DNA-glycosylase) and BER (base excision repair)-mediated pathways
DNA methylation is also present in insects, but at lower levels than in mammals and plants, e.g., less than 1% of CpGs are methylated in honey bees and less than 0.2% in Florida Carpenter ants. In contrast to mammals, methylated cytosines are mainly enriched in gene bodies in insects [14,15]. While DNA methylation has been mainly studied in eusocial insects [14,15], a recent study on the presence of DNA methylation in members of the genus Drosophila established the presence of DNA methylation in the common model organism D. melanogaster and eleven other Drosophila species at low but significant levels [16]. Notably, D. melanogaster was shown to have the lowest level of 5mC amongst the 12 analysed Drosophila genomes that ranged between 0.001% (D. melanogaster) and 0.093% (D. persimilis) 5mC nucleotides per cytosine nucleotides [16].
In D. melanogaster, the presence and functional role of DNA 5-methylcytosine (5mC) modification has been controversial. While some studies have reported the presence of 5mC in the D. melanogaster genome at a low percentage in all stages [17], others have reported the presence of 5mC primarily at early stages of development [18,19], and yet others have reported the complete absence of DNA methylation in 0–2 h old D. melanogaster embryos [20]. These variations appear to be due to the fact that older studies relied on bisulphite sequencing, which is not sensitive enough to detect lower levels of DNA methylation. Particularly in very early embryos (0–2 h), DNA methylation is likely to be extremely low due to the absence of maternal contribution of methylating and demethylating enzymes. Recent studies that employed more sensitive techniques than bisulphite sequencing such as liquid chromatography selective reaction monitoring (LC-SRM) or ultra-high performance liquid chromatography/triple quadrupole mass spectrometry analysis (UHPLC/MS/MS), have reported low but significant levels of 5mC during different developmental stages and in adult flies [21]. Higher levels of 5mC nucleotides per cytosine nucleotides are seen in early stages of development (embryo stage 12–16: 0.026%, larval: 0.025%) and subsequently decrease significantly during pupal stages (0.001–0.002%) and in adult flies (0.001%) [16].
Interestingly, like most dipterans, Drosophila are known to lack the maintenance DNA-methyltransferase 1 (DNMT1), as well as DNA-methyltransferase 3A and B (DNMT3A/B) that target unmethylated cytosine for de novo methylation and only possess a single homolog of DNA-methyltransferase 2 (DNMT2) [15,22]. Similar to mammalian DNMT2, D. melanogaster Dnmt2 is primarily an RNA methyl-transferase that methylates several tRNAs [7,23]. Dnmt2-mediated methylation can protect tRNAs against stress-induced ribonuclease cleavage. This indicates a possible role for Dnmt2 enzymes in regulating the biogenesis of tRNA-derived small RNAs which in turn can act to downregulate target mRNAs [7]. In line with that, loss-of-function mutants of D. melanogaster Dnmt2 have no obvious phenotype under standard laboratory conditions [23], but show reduced viability under stress conditions [7]. In fact, the genome-wide methylcytosine patterns in two different Dnmt2 mutants were unchanged compared to wild-type flies, raising the question whether D. melanogaster harbours a yet unidentified DNA methyltransferase enzyme [19]. In conclusion, D. melanogaster DNA contains 5mC at low levels throughout all developmental stages yet its biological function and the methylating enzyme involved remain elusive. However, more recent findings indicate that the functional role of 5mC DNA methylation might be replaced by other epigenetic marks such as 5mC RNA methylation and 6mA DNA methylation in D. melanogaster [4,9].
The discovery of new epigenetic marks in Drosophila
In 2015, a study by Zhang et al. detected for the first time 6mA in the Drosophila genome using dot blot and UHPLC-MRM-MS/MS assays. 6mA levels were found to be highest in early stages of Drosophila embryogenesis (0.75–2 h/~0.07% 6 mA/dA), then drop throughout later embryonic stages (4–16 h/~0.001% 6 mA/dA), but could still be detected in adult tissues including brain and ovary. Moreover, Zhang et al. showed that nuclear protein extracts from Drosophila embryos displayed a dynamic 6mA demethylating activity that peaked at 6 h of embryogenesis. This 6mA demethylating activity was lost in dTetnull mutants. An in vitro assay using purified dTet catalytic domain (aa1657-2918) and a catalytic-impaired version where the two Fe(II)-binding sites H1948 and D1950 were mutated to Y and A, respectively, showed that the dTet catalytic domain is capable of direct 6mA demethylation; this 6mA demethylation activity was 4–5 times reduced upon using the catalytic mutant version. Importantly, the same dTet catalytic domain was found to be capable of demethylating 5mC on DNA in vitro, which shows its conserved function across vertebrates and invertebrates alike [4]. The 6mA demethylating activity of dTet is somehow puzzling as sequence alignments of dTet and mammalian TETs revealed that the active-site residues of dTet are absolutely conserved to mammalian TET active-site residues involved in interactions with the pyrimidine ring of 5mC (Asn and His) [24]. It is therefore surprising that the same active site could accommodate the larger purine ring of adenine to allow for 6mA demethylation. In fact, dTet belongs to the TET/kinetoplastid base J-binding protein (JBP) family that shares sequence similarities as well as enzymatic features with homologs present in all domains of life (from virus to human) [25]. This well-conserved enzyme family can oxidize 5-methylpyrimidines, but so far only dTet has been linked to oxidizing methylpurines [4].
Recently, two additional studies have detected 6mA in the Drosophila genome: one used techniques similar to the ones previously published and detected 0.0026% 6 mA/dA in fly brains [26] and another used Nanopore sequencing [27]. Both found 6mA enriched at intergenic regions, within introns, and at simple repeats [26,27]. Analysis of the sequence motifs at which 6mA is preferentially found showed that these are also binding sites for transcriptional activators involved in development including Bicoid and Caudal, which are required for regulating anterior/posterior patterning during development [27].
The DNA modification 6mA has well-described functions in prokaryotes particularly in restriction-modification systems important for host-defences [28], DNA replication and repair [29], as well as in gene expression [30]. In eukaryotes, 6mA has been associated with diverse functions including nucleosome positioning in ciliates [31], active gene transcription in algae, worms, and humans [2,32,33], and transposon expression in flies [4]. This diversity, therefore, suggests that this modification has evolved species-specific functional roles. In Drosophila, 6mA has been shown to be important for regulating the brain-specific expression of a group of genes involved in neurodevelopment and neuronal functions by coordinating with the Trithorax and Polycomb system [26] as well as for regulating transposon expression in ovaries of adult flies [4]. Notably, genome wide-mapping of 6mA-containing genes in adult females found that 6mA-containing genes display a high tissue-specificity in several tissues besides the brain, indicating that this modification may play a more general role in regulating the tissue-specific expression of genes across many tissues and during development [27]. Therefore, 6mA in eukaryotic genomes appears to be a versatile mark whose effect is not only species, but also context-dependent. In addition to other pertinent factors [34], in certain tissues, such as the brain, or developmental time points, the deposition of 6mA may trigger different effects on gene expression depending on factors that may include readers and downstream pathways that are involved.
Besides its role in 6mA demethylation, dTet was also found to catalyse the hydroxymethylation of 5mrC into 5hmrC. This discovery fuelled further interest in the use of fly to study RNA modifications since 5hmrC had not been previously described in mammals. 5hmrC was preferentially found on polyadenylated RNAs and transcriptome-wide mapping revealed its enrichment in exons and in a specific sequence context (UCCUC repeats). In addition, it was enriched at genes involved in developmental processes and embryogenesis [9]. Although data on the functional role of 5hmrC is still lacking, this study has provided a hint that 5hmrC may favour translation of mRNA transcripts as ribosome load was increased on 5hmrC-containing RNAs. Another study in mammals indicated a possible role for 5mrC in regulating mRNA export from the nucleus [35]. Further studies are required to determine the biological outcome of this RNA modification. It will then be possible to have a better understanding of the role of dTet in demethylating 6mA versus 5mrC and whether these two marks are important at distinct developmental stages or tissues.
DNA 6-methyladenine and RNA 5-methylcytosine in mammals
After 6mA and 5hmrC were discovered in flies, mammalian tissues were closely probed for their presence, as summarized in (Figure 1a, Table 1). Interestingly, in a mouse study, 6mA was shown to be a highly adaptive mark as its presence was affected by environmental conditions. When mice were exposed to chronic restraint stress, 6mA levels increased significantly in the brain [3]. This was associated with decreased expression of transposons, which is in line with data previously reported in flies [4]. Transposons contribute to genomic instability and may lead to gene mutations due to insertions at random loci. Accordingly, various transposable elements are associated with diseases such as cancer, in which transposon promoters are demethylated (Reviewed in [36]). This highlights the possible relevance of 6mA, and therefore dTet, in regulating transposon expression at an appropriate level.
Table 1.
The function of 6mA and 5hmrC modification in different multicellular organisms
| Modification | Multicellular organism | Function | Correlation with gene expression | Reference |
|---|---|---|---|---|
| 6mA | A. thaliana | protein coding gene regulation | activation | Liang et al. 2018 |
| B. mori | regulates cell cycle progression | repression | Wang et al. 2018 | |
| C. elegans | gene regulation, regulation of stress response, transgenerational inheritance | activation |
Greer et al. 2015 Ma et al. 2019 |
|
| D. melanogaster | regulates transposon expression in ovary regulates neuronal gene expression in brains regulates transcription in whole fly |
activation repression repression |
Zhang et al. 2015 Yao et al. 2018 Shah et al. 2019 |
|
| D. rerio and S. scrofa | involved in embryogenesis, activation of repetitive elements | activation | Liu et al. 2016 | |
| H. sapiens | prevents tumorigenesis | activation | Xiao et al. 2018 | |
| M. musculus | regulates expression of neuronal genes and LINE transposon expression in stress condition | repression | Yao et al. 2017 | |
| O. sativa | gene regulation, repression of transposable element-related genes | gene-dependent | Zhou et al. 2018 | |
| T. thermophile | contributes to nucleosome positioning | gene-dependent | Wang et al. 2017 | |
| X. laevis | regulates transcription, nucleic acid binding, metabolic processes, cell adhesion and ATP binding activity | N/A | Koziol et al. 2016 | |
| 5hmrC | D. melanogaster | regulates embryogenesis/development | gene-dependent | Delatte et al. 2016 |
| M. musculus | decrease detected in model of Parkinson’s disease | N/A | Miao et al. 2016 | |
| M. musculus and H. sapiens | regulates gene expression | N/A | Fu et al. 2014 |
Abbreviations
5mC: 5-methylcytosine
5hmC: 5-hydroxylmethylcytosine
TET: Ten-eleven Translocation
DNMT: DNA methyltransferase
Similar to the finding in Drosophila brains [26], 6mA enrichment was also correlated with altered expression of important neuronal genes in mice [3]. Interestingly, many 6mA dynamic genes overlapped with genes associated with depression, autism, and schizophrenia, further consolidating the link between 6mA regulation and disease [3]. This was also confirmed in human cancer tissues, upon the discovery that a decrease in 6mA levels may be involved in tumorigenesis [33]. In another paper, 6mA was found to be enriched in primary human glioblastoma cells and brain tissues [37]. This was associated with repression of tumour suppressor genes such as Cyclin-dependent kinase inhibitor 3 (CDKN3) and Ras association domain family member 2 (RASSF2). Due to these conflicting findings on whether global 6mA is increased or decreased in glioblastoma, additional studies on glioblastoma patient samples are required to elucidate the involvement of 6mA in glioblastoma and other cancers.
5hmrC was found to be present in various human cell lines as well as mouse and human tissues with the highest levels of 5hmrC detected in brain and heart tissues [38] (Figure 2b). Levels were decreased upon triple knockdown of TETs 1, 2, and 3 [38]. In the mouse brain, 5hmrC levels were highest in the cerebellum, hippocampus, and brain stem [39]. In a mouse model for Parkinson’s disease, 5hmrC levels were significantly reduced in the substantia nigra and striatum, two regions that are highly implicated in the pathology of the disease [39]. TET proteins also play an important role in zygotic epigenetic reprogramming, pluripotent stem cell differentiation and haematopoiesis (Figure 3) [11]. Moreover, unpublished data from our group also revealed dTet expression in the larval and pupal fat body (an organ involved in immune response) [modEncode [40]; Shirinian lab, unpublished]; however, whether dTet plays a role in Drosophila haematopoiesis remains to be clarified.
Figure 2.
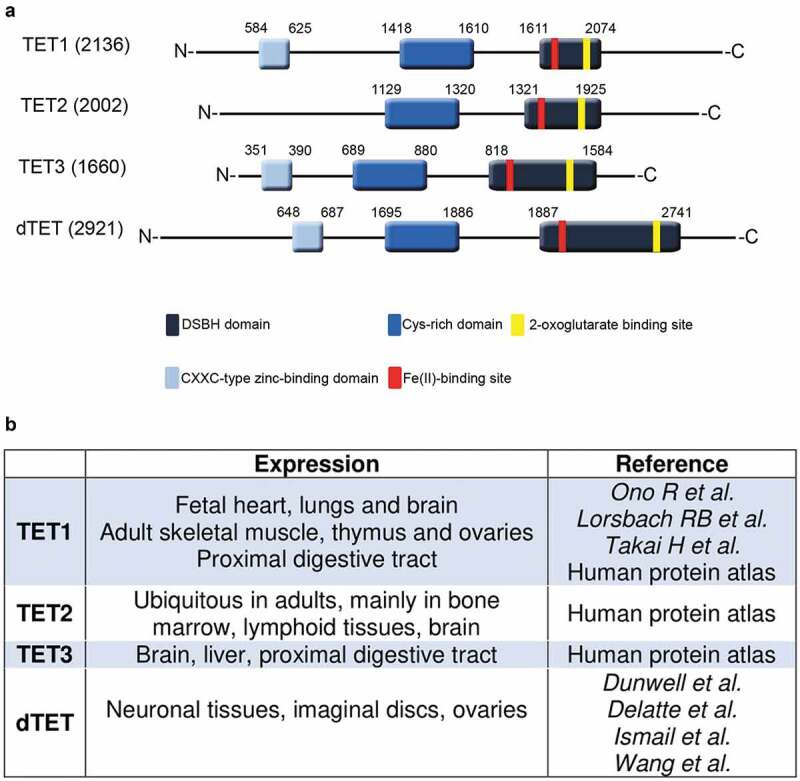
Comparison of human and D. melanogaster TET protein structure and expression. (a). Three TET proteins are found in humans in comparison to one TET (dTet) homologue in D. melanogaster. All share a conserved cysteine-rich domain (dark blue colour) and a DSBH (Double Stranded Beta-Helix) domain (dark grey colour). TET2 lacks the CXXC DNA binding domain (light blue colour). The Fe(II)-binding site (red colour) and 2-oxoglutarate binding site (yellow colour) are both located within the catalytic domain. (b). An overview of TET expressing tissues in humans and D. melanogaster.
Figure 3.
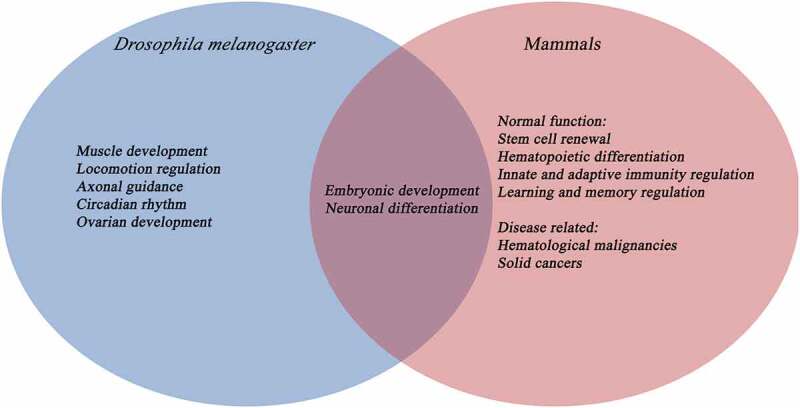
Comparison between the function of TET proteins in D. melanogaster and mammals
To decipher the functional role of 6mA and 5hmrC on a molecular level, a deep understanding of the pathways that are involved is essential, especially on ‘readers’ which are molecules responsible for interpreting these marks and triggering downstream effectors that result in a change in gene expression. Many readers of 5hmC on DNA have been investigated. Some well-characterized mammalian readers include Ubiquitin-like with PHD and Ring Finger Domains 2 (UHRF2) and Methyl-CpG-binding Protein 2 (MeCP2) [41,42]. Whether the readers for 5hmC on DNA and RNA are the same is yet to be determined. Furthermore, only a few 6mA readers have been identified, such as the YT521-B homology (YTH) domain proteins, which are known to regulate RNA signalling pathways [43]. In the fly, readers of 5hmC have not been identified yet. However, a conserved YT521 protein has been shown to be an RNA 6mA reader [44]. Further studies into the readers, writers, and erasers of these novel marks will contribute to our overall understanding of the pathways that are activated upon demethylation of 6mA or 5hmrC.
Structural basis of Drosophila Tet protein
The Drosophila genome contains a single Tet gene that encodes six annotated transcripts. Of these six transcripts, two are short and only contain the catalytic double-stranded beta helix (DSBH) domain, while the other four are longer and contain an additional N-terminal CXXC zinc finger domain that has putative DNA binding activity. dTet is evolutionarily conserved to the three mammalian TET proteins, particularly the amino acid sequence in the functional domains (Figure 2a). Three-dimensional modelling and comparison between human TET1, TET2 and TET3 and the dTet catalytic domain showed that dTet is most closely related to hTET3 [45], the only mammalian TET protein that is essential for development and is highly expressed in the nervous system during development [46,47]. Comparably, dTet is also required for normal development and viability.
Two studies addressed the functional requirements of the conserved domains of dTet by generating domain-deletion alleles. The first study used the CRISPR/Cas system to generate an N-terminal deletion that lacks the CXXC domain and a C-terminal deletion that removes the DSBH catalytic domain. While the CXXC domain is dispensable for the role of dTet during development and is not required for catalytic activity, the catalytic domain of dTet is required for development and a trans-heterozygous mutant showed a significant increase in the 6mA mark in its genomic DNA [4]. Conversely, a second study that used FRT-mediated site-specific recombination of piggyBac transposons to generate truncated versions of the dTet protein found that both N- and C-terminally truncated dTet led to pupal lethality, either homozygous or trans-heterozygous over dTetnull. Moreover, both, N- and C-terminally truncated dTet mutant larvae, displayed clear defects in locomotion [48]. Together, this suggests that the catalytic domain is essential for the role of dTet in development; however, the conflicting results about the necessity of the CXXC domain requires further investigation and might be due to the fact that both studies used deletion constructs of different length.
Dynamic expression of Drosophila Tet during development
dTet expression is dynamic and tightly regulated during development. In situ staining and mRNA seq data have shown that dTet expression is absent in early embryos (0–2 h/no maternal contribution) and is only detected from 3 h post-fertilization (stage 7) onwards. dTet is first expressed in ectoderm tissues, the hindgut, and ventral nerve cord (VNC) (stages 7 to 10). At 6–8 h post-fertilization expression of dTet peaks (~stage 11 to 12). At this stage, dTet is observed ubiquitously at faint levels and at higher levels in the central nervous system (CNS). Expression of dTet decreases by stages 13 to 16 and becomes limited to the nervous system [modEncode 40,48,49]. Similar embryonic expression profiles have been observed with the endogenous dTet-GFP reporter [48] and by western blot using a dTet-specific antibody [4]. After embryogenesis, dTet is mainly expressed in the CNS of L3 wandering larvae (particularly in neurons, the optic lobe, and midline glia [50]); however, it is also expressed at lower levels in imaginal discs and the carcass [9]. In pupal and adult stages, there is less dTet expression and it is mainly concentrated in the head and CNS compared to other tissues such as the ovary [4,9,45]. Taken together, dTet expression is tightly controlled and the protein is most abundantly expressed in the nervous system throughout development, but dTet expression is also required in other tissues such as the ovaries and somatic muscle precursors at specific developmental time points [4,48].
Functional role of Drosophila Tet during development
One of the advantages of the Drosophila system is the lack of genetic redundancy compared to other model organisms. D. melanogaster has only a single Tet gene as opposed to three TET genes that have overlapping functions in mammals. Therefore, it is easier and more straight-forward to generate dTet knockout (dTetnull) flies than for example TET1/2/3 knockout mice and study resulting mutant phenotypes. Currently, there are six reported partial or full dTetnull alleles, which differ in size and location of the underlying genomic deletion and were generated with different genetic techniques including CRISPR/Cas and transgenic transposon insertion. Notably, the exact time point that dTetnull-animals die differs slightly depending on the alleles used to generate dTetnull-animals [4,9]. Differences in survival rates could either be due to secondary (off-target) mutations on the more severe dTetnull allele or to an only partial loss of function mutations on less severe dTetnull alleles. Despite these slight variations in survival, all dTetnull flies died within 2 days of eclosion and displayed a strong locomotion phenotype that was consistently observed in all mutant alleles [48]. Here, we will summarize some of the important roles of dTet.
Role of dTet in brain and muscle precursors
One of the most striking phenotypes that dTetnull larvae and adult escapers display is impaired locomotion. In motility assays dTetnull and dTet-depleted larvae were shown to exhibit severe loss of motility and body wall contractions indicating that dTet is required for normal locomotion [48,50]. Locomotion behaviours are coordinated by neurons, motor neurons, neuromuscular junctions, and muscles, which seem to be affected to different degrees in dTetnull larvae.
Muscle-specific knockdown of dTet resulted in larval locomotion defects similar to those observed in dTetnull animals suggesting that dTet is required in somatic muscle tissue for normal locomotion. Interestingly, dTet was shown to be essential in somatic muscles at early stages of development (embryogenesis) and dispensable at the larval stages [48].
dTetnull larvae were reported to show no significant changes in morphology or number of boutons in the neuromuscular junctions (NMJs) of motor neurons terminating on the somatic musculature. In line with that, motor neuron-specific knockdown of dTet did not result in a locomotion phenotype [48]. In contrast, we found that dTetnull larvae (Mi(MIC)TetMI03920/Df(3 L)Exel6091) display small, but significant changes in sarcomere organization (increased number of split muscles and unorganized sarcomeric structure) (Shirinian lab, unpublished). While NMJ branching was not affected, there was a significant decrease in muscle nuclei and an increase in bouton numbers. However, these changes do not fully explain the severe locomotion phenotype observed, indicating that the requirement for dTet in locomotion is not primarily in NMJs.
dTet expression is highest in the nervous system throughout development, further stressing the importance of dTet for brain development. While the embryonic nervous system and third instar larval brains of dTetnull animals show no major difference in organization or morphology compared to wild-type, neuron- or midline glia-specific depletion of dTet resulted in severe locomotion defects [48] and additional phenotypes in mushroom body organization (neuronal) [26] or axon patterning (midline glial) [50], respectively. Neuron-specific knockdown of dTet showed that besides its function in locomotion, dTet also plays a role in maintaining the normal circadian rhythm in a subgroup of neurons called pigment-dispersing factor (PDF) neurons by controlling the development of precursors into functional PDF neurons [48]. Moreover, dTet expression was detected in midline glia cells in the late embryonic CNS and in third instar larval VNCs [50]. Midline glia play an important role in axon guidance during brain development and dTet depletion with either ubiquitous or midline glia-specific drivers resulted in impaired locomotion as well as disruption of axons and midline commissure organization in third instar larval VNCs suggesting that dTet function in midline glia contributes to normal locomotion [50]. In addition, neuronal knockdown of dTet resulted in aberrant midline cleft crossings of mushroom body β-lobes in adult flies [26].
Interestingly, dTet depletion in flies leads to downregulation of genes involved in behaviour, learning, memory, and neuronal differentiation accompanied by an increase of 6 mA in the same genomic regions [26]. Similarly, gene expression analysis by RNA-seq upon dTet depletion in a fly neuronal cell line (BG3 C2) showed that particular genes involved in processes such as neuron development, neuron differentiation, axon guidance, and axogenesis are downregulated, while simultaneously displaying intragenic gain-of-6mA regions [26]. In line with that, genes involved in axon guidance and axonogenesis such as slit and prospero have been demonstrated to be downregulated in dTet-depleted third instar brains [50]. Moreover, dTet has been suggested to work cooperatively with the Trithorax/Polycomb system via direct interaction with the Trithorax-related protein Will die slowly (Wds) in fly neurons to activate transcription of a group of neuron-specific genes (involved in axonogenesis and neuronal development and differentiation) by removing intragenic 6mA. Depletion of dTet resulted in a reduction of Wds binding and accumulation of 6mA. This 6mA accumulation was shown to coincide with an increase in Polycomb (Pc) binding suggesting that recruitment of Polycomb proteins implements transcriptional repression on these loci [26].
Role of dTet in ovaries
dTetnull mutants display a strong phenotype in larval and adult ovaries [4,48] despite low dTet expression in this organ [9]. While low levels of dTet expression were detected in germaria and egg chambers of Drosophila ovaries particularly in the nucleus of germ cells when using a dTet specific antibody [4], an endogenous dTet-GFP reporter did not show expression in adult fly ovaries or in somatic and germline cells, and was absent in embryonic gonads [48]. However, weak nuclear expression was detected in the terminal filament cells in gonads of third instar larvae, which might explain the disrupted ovary phenotype present in dTetnull animals. dTetnull ovaries showed an increase in the average of spectrosome-containing germ cells (GSC-like cells) as compared to wild-type ovaries [4]. Moreover, dTet overexpression using a germ-cell-specific driver resulted in a significant loss of germ cells, therefore, indicating that dTet plays a role in promoting early germ cell differentiation [4]. In contrast, Wang et al. reported that ovaries in dTetnull larvae and pharate adults showed disrupted organization without loss of germ cells. In fact, ovarioles failed to separate and early egg chambers and somatic parts of the ovary were not detected with the exception of a few stage 8 egg chambers [48]. Furthermore, Wang et al. showed that induction of germ line clones during embryonic and not third instar larval stage resulted in the reduction of the number of dTetnull egg chambers as compared to controls, implicating the requirement of dTet in oogenesis during embryonic and early larval stages [48].
Intriguingly, dTet mutant ovaries showed a 10-fold increase in 6mA levels. In wild-type ovaries, high levels of 6mA were detected in the less differentiated germarium cells, including the germ and somatic cells, which then dropped throughout development and in the more differentiated egg compartments. While nuclear extract from wild-type ovaries had a strong demethylation activity for 6mA, nuclear extract from dTetnull ovaries displayed very low levels of 6mA demethylation activity in in vitro assays. Addition of purified dTet catalytic domain (1657–2918aa) restored the demethylation activity, whereas addition of dTet catalytic mutant domain (1657–2918aaY1948A1950) did not, implicating the catalytic domain of dTet in direct demethylation of 6mA [4]. This 6mA demethylation activity is correlated with suppression of transposon expression and hence gene regulation. If this activating effect of 6mA on transposon expression is a germline-specific phenomenon in Drosophila or can also be observed in somatic cells remains to be clarified. Finally, the expression pattern differences seen between the two studies could be due to some unspecific anti-dTet antibody binding, since the dTet-GFP line expression pattern reported by Wang et al. is also in line with data from modEncode [40].
Conclusion and outlook
In this review, we discussed recent findings on the role of DNA/RNA demethylation by dTet and the impact of the novel 6mA DNA and 5hmC RNA marks on development. These insights highlight the need for more studies that can contribute to our understanding of how these two modifications act during development in the fly and beyond.
The physiological function of 6mA and its effect on gene expression needs to be further investigated. Existing data on the functional role and distribution pattern of 6mA in eukaryotes point at species-specific roles, since neither methylation patterns nor biological roles appear to be strictly conserved [2,4,26,32,33]. Moreover, data from Drosophila indicate that even within the same organism 6mA might have different, tissue-specific functions [4,26]. Future studies on detailed molecular mechanisms of 6mA-mediated regulation of gene and transposon expression are required and will help to understand how the same DNA modification can result in diverse species- and tissue-specific outputs. Identifying specific 6mA writers, erasers, and readers in different eukaryotes will be an important step in further unravelling the role of this mark. While writers and erasers have been identified for several organisms including worms [2] and humans [33], data on readers of 6mA in other eukaryotes are still lacking. Interestingly, the reported genomic 6mA demethylase ALKBH1 in human and mouse ALKBH1 are highly homologous [33] and belong, together with NMAD-1, the 6mA demethylase identified in C. elegans [2], to the AlkB family of dioxygenases, a class of DNA/RNA repair enzymes that remove alkyl adducts such as m6A and m3C from nucleobases by oxidative dealkylation [33,51]. Conversely, the reported 6mA demethylase in Drosophila belongs to the TET/JBP family and appears to have extended its substrate specificity in flies. One intriguing question that remains to be tackled is how dTet is targeted to mediate selective oxidation for its substrates (RNA versus DNA, respectively, 5mrC versus 6mA) and whether this activity spectrum is fly-specific. Studies have shown that mammalian TET enzymes are promiscuous and can oxidize 5mC and 5mrC in vitro [38]. Recently, a detailed biochemical analysis of the catalytic domain of human TET2 found that TET2 is most proficient at dsDNA oxidation, discriminates strongly against dsRNA, but tolerates ssDNA, ssRNA as well as hybrid substrates [52]. Therefore, the reported 5mrC demethylating activity [9] of the well-conserved dTet in the absence of abundant DNA methylation in the fly is no surprise; it is more difficult to reconcile how dTet would have managed to broaden its substrate specificity from 5mC to 6mA in DNA without changes to the active site of the catalytic domain. Structural studies of dTet’s catalytic domain in complex with its substrates including 6mA and 5mrC will be necessary to resolve this puzzling question. Whether TET enzymes have evolved to support diverse biological functions in different organisms is still elusive and could be elegantly addressed by assessing its proficiency towards different substrates in various species. An additional open question is which methyltransferases are catalysing 6mA and 5mrC in Drosophila. Further assessment on the role of TET substrates and oxidized TET derivatives (5hmc, 5fc, 5caC) as independent epigenetic marks will help us to understand the stability and the dynamics of these marks in different pathologies such as cancers, neurological, and autoimmune diseases. In this regard, writers and readers of the DNA/RNA marks as well as genes that generate the cofactors for TET such as the citric acid cycle enzymes, isocitrate dehydrogenase 1 and 2 (IDH1 and 2), should be thoroughly investigated. Furthermore, it has been suggested that dTet may affect gene expression independently of its catalytic function [45] for example, through interacting proteins. Intriguingly, mammalian TET proteins have been shown to interact with the O-linked N-acetylglucosamine (O-GlcNAc) transferase (Ogt), which also catalyses histone modifications thus linking TETs to other epigenetic control mechanisms besides DNA demethylation [53]. Therefore, studying the non-catalytic role of TET will enrich our understanding of TET proteins in health and disease.**** [54–58].
Acknowledgments
We thank Elias A. Rahal for critical reading of the review.
Funding Statement
This work is funded by Medical Practice Plan (MPP) at the American University of Beirut. JI and AA are recipients of CNRS Conseil National de la Recherche Scientifique (CNRS)award.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- [1].Flavahan WA, Gaskell E, Bernstein BE.. Epigenetic plasticity and the hallmarks of cancer. Science. 2017;357(6348):eaal2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Greer EL, Blanco MA, Gu L, et al. DNA methylation on N6-adenine in C. elegans. Cell. 2015;161(4):868–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Yao B, Cheng Y, Wang Z, et al. DNA N6-methyladenine is dynamically regulated in the mouse brain following environmental stress. Nat Commun. 2017;8. DOI: 10.1038/s41467-017-01195-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Zhang G, Huang H, Liu D, et al. N6-methyladenine DNA modification in Drosophila. Cell. 2015;161(4):893–906. [DOI] [PubMed] [Google Scholar]
- [5].Boccaletto P, Machnicka MA, Purta E, et al. MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic Acids Res. 2018;46(D1):D303–D307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Frye M, Jaffrey SR, Pan T, et al. RNA modifications: what have we learned and where are we headed? Nat Rev Genet. 2016;17(6):365–372. [DOI] [PubMed] [Google Scholar]
- [7].Schaefer M, Pollex T, Hanna K, et al. RNA methylation by Dnmt2 protects transfer RNAs against stress-induced cleavage. Genes Dev. 2010;24(15):1590–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Squires JE, Patel HR, Nousch M, et al. Widespread occurrence of 5-methylcytosine in human coding and non-coding RNA. Nucleic Acids Res. 2012;40(11):5023–5033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Delatte B, Wang F, Ngoc LV, et al. RNA biochemistry. Transcriptome-wide distribution and function of RNA hydroxymethylcytosine. Science. 2016;351(6270):282–285. [DOI] [PubMed] [Google Scholar]
- [10].Li E, Zhang Y.. DNA methylation in mammals. Cold Spring Harb Perspect Biol. 2014;6(5):a019133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Wu H, Zhang Y. Reversing DNA methylation: mechanisms, genomics, and biological functions. Cell. 2014;156(1–2):45–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Monk M, Boubelik M, Lehnert S. Temporal and regional changes in DNA methylation in the embryonic, extraembryonic and germ cell lineages during mouse embryo development. Development. 1987;99(3):371–382. [DOI] [PubMed] [Google Scholar]
- [13].Edwards JR, Yarychkivska O, Boulard M, et al. DNA methylation and DNA methyltransferases. Epigenetics Chromatin. 2017;10:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Bewick AJ, Vogel KJ, Moore AJ, et al. Evolution of DNA methylation across insects. Mol Biol Evol. 2017;34(3):654–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Yan H, Bonasio R, Simola DF, et al. DNA methylation in social insects: how epigenetics can control behavior and longevity. Annu Rev Entomol. 2015;60:435–452. [DOI] [PubMed] [Google Scholar]
- [16].Deshmukh S, Ponnaluri VC, Dai N, et al. Levels of DNA cytosine methylation in the Drosophila genome. PeerJ. 2018;6:e5119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Gowher H, Leismann O, Jeltsch A. DNA of Drosophila melanogaster contains 5-methylcytosine. Embo J. 2000;19(24):6918–6923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Lyko F, Ramsahoye BH, Jaenisch R. DNA methylation in Drosophila melanogaster. Nature. 2000;408(6812):538–540. [DOI] [PubMed] [Google Scholar]
- [19].Takayama S, Dhahbi J, Roberts A, et al. Genome methylation in D. melanogaster is found at specific short motifs and is independent of DNMT2 activity. Genome Res. 2014;24(5):821–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Raddatz G, Guzzardo PM, Olova N, et al. Dnmt2-dependent methylomes lack defined DNA methylation patterns. Proc Natl Acad Sci U S A. 2013;110(21):8627–8631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Capuano F, Mülleder M, Kok R, et al. Cytosine DNA methylation is found in Drosophila melanogaster but absent in Saccharomyces cerevisiae, Schizosaccharomyces pombe, and other yeast species. Anal Chem. 2014;86(8):3697–3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Glastad KM, Hunt BG, Yi SV, et al. Epigenetic inheritance and genome regulation: is DNA methylation linked to ploidy in haplodiploid insects? Proc R Inst Soc B-Biol Sci. 2014;281(1785). DOI: 10.1098/rspb.2014.0411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Goll MG, Kirpekar F, Maggert KA, et al. Methylation of tRNAAsp by the DNA methyltransferase homolog Dnmt2. Science. 2006;311(5759):395–398. [DOI] [PubMed] [Google Scholar]
- [24].Parker MJ, Weigele PR, Saleh L. Insights into the biochemistry, evolution, and biotechnological applications of the ten-eleven translocation (TET) enzymes. Biochemistry. 2019;58(6):450–467. [DOI] [PubMed] [Google Scholar]
- [25].Iyer LM, Tahiliani M, Rao A, et al. Prediction of novel families of enzymes involved in oxidative and other complex modifications of bases in nucleic acids. Cell Cycle. 2009;8(11):1698–1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Yao B, Li Y, Wang Z, et al. Active N(6)-methyladenine Demethylation by DMAD regulates gene expression by coordinating with polycomb protein in neurons. Mol Cell. 2018;71(5):848–857 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Shah K, Cao W, Ellison CE. Adenine methylation in Drosophila is associated with the tissue-specific expression of developmental and regulatory genes. G3 (Bethesda). 2019;9(6):1893–1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Naito T, Kusano K, Kobayashi I. Selfish behavior of restriction-modification systems. Science. 1995;267(5199):897–899. [DOI] [PubMed] [Google Scholar]
- [29].Reisenauer A, Kahng LS, McCollum S, et al. Bacterial DNA methylation: a cell cycle regulator? J Bacteriol. 1999;181(17):5135–5139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Lobner-Olesen A, Skovgaard O, Marinus MG. Dam methylation: coordinating cellular processes. Curr Opin Microbiol. 2005;8(2):154–160. [DOI] [PubMed] [Google Scholar]
- [31].Luo GZ, Hao Z, Luo L, et al. N(6)-methyldeoxyadenosine directs nucleosome positioning in tetrahymena DNA. Genome Biol. 2018;19(1):200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Fu Y, Luo GZ, Chen K, et al. N6-methyldeoxyadenosine marks active transcription start sites in chlamydomonas. Cell. 2015;161(4):879–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Xiao CL, Zhu S, He M, et al. N(6)-methyladenine DNA modification in the human genome. Mol Cell. 2018;71(2):306–318 e7. [DOI] [PubMed] [Google Scholar]
- [34].Fontebasso AM, Shirinian M, Khuong-Quang DA, et al. Non-random aneuploidy specifies subgroups of pilocytic astrocytoma and correlates with older age. Oncotarget. 2015;6(31):31844–31856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Yang X, Yang Y, Sun BF, et al. 5-methylcytosine promotes mRNA export - NSUN2 as the methyltransferase and ALYREF as an m(5)C reader. Cell Res. 2017;27(5):606–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Ayarpadikannan S, Kim HS. The impact of transposable elements in genome evolution and genetic instability and their implications in various diseases. Genomics Inform. 2014;12(3):98–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Xie Q, Wu TP, Gimple RC, et al. N(6)-methyladenine DNA modification in glioblastoma. Cell. 2018;175(5):1228–1243 e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Fu L, Guerrero CR, Zhong N, et al. Tet-mediated formation of 5-hydroxymethylcytosine in RNA. J Am Chem Soc. 2014;136(33):11582–11585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Miao Z, Xin N, Wei B, et al. 5-hydroxymethylcytosine is detected in RNA from mouse brain tissues. Brain Res. 2016;1642:546–552. [DOI] [PubMed] [Google Scholar]
- [40].Celniker SE, Dillon LA, Gerstein MB, et al. Unlocking the secrets of the genome. Nature. 2009;459(7249):927–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Chen R, Zhang Q, Duan X, et al. The 5-hydroxymethylcytosine (5hmC) reader UHRF2 is required for normal levels of 5hmC in mouse adult brain and spatial learning and memory. J Biol Chem. 2017;292(11):4533–4543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Mellen M, Ayata P, Dewell S, et al. MeCP2 binds to 5hmC enriched within active genes and accessible chromatin in the nervous system. Cell. 2012;151(7):1417–1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Liao S, Sun H, Xu C. YTH domain: A family of N(6)-methyladenosine (m(6)A) readers. Genomics Proteomics Bioinformatics. 2018;16(2):99–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Guo J, Tang HW, Li J, et al. Xio is a component of the Drosophila sex determination pathway and RNA N(6)-methyladenosine methyltransferase complex. Proc Natl Acad Sci U S A. 2018;115(14):3674–3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Dunwell TL, McGuffin LJ, Dunwell JM, et al. The mysterious presence of a 5-methylcytosine oxidase in the Drosophila genome: possible explanations. Cell Cycle. 2013;12(21):3357–3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Gu TP, Guo F, Yang H, et al. The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature. 2011;477(7366):606–610. [DOI] [PubMed] [Google Scholar]
- [47].Hahn MA, Qiu R, Wu X, et al. Dynamics of 5-hydroxymethylcytosine and chromatin marks in mammalian neurogenesis. Cell Rep. 2013;3(2):291–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Wang F, Minakhina S, Tran H, et al. Tet protein function during Drosophila development. PLoS One. 2018;13(1):e0190367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Brody T, Stivers C, Nagle J, et al. Identification of novel Drosophila neural precursor genes using a differential embryonic head cDNA screen. Mech Dev. 2002;113(1):41–59. [DOI] [PubMed] [Google Scholar]
- [50].Ismail JN, Badini S, Frey F, et al. Drosophila Tet is expressed in midline glia and is required for proper axonal development. Front Cell Neurosci. 2019;13:252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Fedeles BI, Singh V, Delaney JC, et al. The AlkB Family of Fe(II)/alpha-ketoglutarate-dependent dioxygenases: repairing nucleic acid alkylation damage and beyond. J Biol Chem. 2015;290(34):20734–20742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].DeNizio JE, Liu MY, Leddin EM, et al. Selectivity and promiscuity in TET-mediated oxidation of 5-methylcytosine in DNA and RNA. Biochemistry. 2019;58(5):411–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Vella P, Scelfo A, Jammula S, et al. Tet proteins connect the O-linked N-acetylglucosamine transferase Ogt to chromatin in embryonic stem cells. Mol Cell. 2013;49(4):645–656. [DOI] [PubMed] [Google Scholar]
- [54].Ono R, Taki T, Taketani T, et al. Lcx, leukemia-associated protein with a cxxc domain, is fused to mll in acute myeloid leukemia with trilineage dysplasia having t(10;11)(q22;q23). Cancer Res. 2002;62:4075–4080. [PubMed] [Google Scholar]
- [55].Lorsbach RB, Moore J, Mathew S, et al. TET1, a member of a novel protein family, is fused to MLL in acute myeloid leukemia containing the t(10;11)(q22;q23). Leukemia. 2003;17(3):637–641. [DOI] [PubMed] [Google Scholar]
- [56]. Takai H, Masuda K, Sato T, et al. 5-hydroxymethylcytosine plays a critical role in glioblastomagenesis by recruiting the chtop-methylosome complex. Cell Rep. 2014;9(1):48–60. [DOI] [PubMed] [Google Scholar]
- [57].Human protein atlas available from http://www.proteinatlas.org.
- [58].Uhlen M, Fagerberg L, BM Hallström, et al. Tissue-based map of the human proteome. Science. 2015;347(6220):1260419. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Human protein atlas available from http://www.proteinatlas.org.


