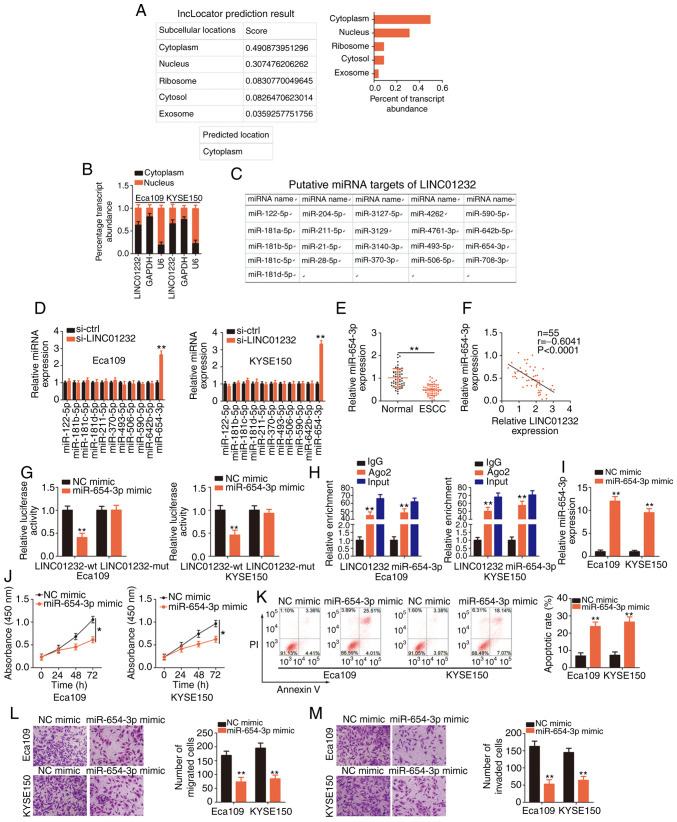
Figure 2.
MicroRNA (miR)-654-3p is sponged by long intergenic non-coding RNA (LINC)01232 and plays an inhibitory role in esophageal squamous cell carcinoma (ESCC). (A) The lncLocator database was used to examine the subcellular location of LINC01232. (B) The subcellular location of LINC01232 was evaluated in cell cytosolic/nuclear fractions isolated from Eca109 and KYSE150 ESCC cells. (C) Potential LINC01232-interacting miRNAs identified through a StarBase 3.0 search. (D) The expression levels of miR-122-5p, miR-181b-5p, miR-181c-5p, miR-181d-5p, miR-211-5p, miR-370-5p, miR-493-5p, miR-506-5p, miR-590-5p, miR-642b-5p and miR-654-3p in LINC01232-deficient Eca109 and KYSE150 cells were detected by reverse transcription-quantitative poly-merase chain reaction (RT-qPCR). (E) RT-qPCR was performed to detect miR-654-3p expression in 55 pairs of ESCC tissues and matched normal tissues. (F) Pearson's correlation coefficient illustrates the correlation between LINC01232 and miR-654-3p in 55 ESCC tissues. (G) Luciferase activity corresponding to LINC01232-wt or LINC01232-mut was detected in Eca109 and KYSE150 cells following transfection with the miR-654-3p mimic or NC mimic. (H) RT-qPCR was conducted to determine the enrichment of LINC01232 and miR-654-3p in Eca109 and KYSE150 cells after RNA immunoprecipitation. (I) Eca109 and KYSE150 cells were transfected with miR-654-3p mimic or NC mimic, and the overexpression efficiency was determined by RT-qPCR. (J-M) CCK-8 assay, flow cytometric analysis, and migration and invasion assays were performed to detect the proliferation, apoptosis, and migration and invasion of miR-654-3p-overexpressing Eca109 and KYSE150 cells. *P<0.05 and **P<0.01 vs. NC mimic or si-ctrl.