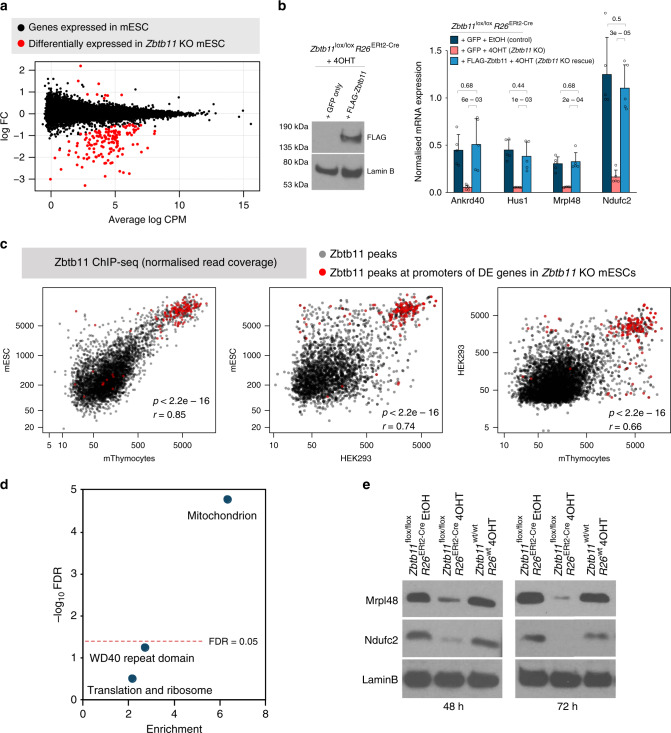
Fig. 3. Zbtb11 supports the expression of a subset of nuclear-encoded mitochondrial proteins.
a Whole-transcriptome analysis by RNA-seq comparing Zbtb11lox/lox Rosa26ERt2-Cre cells treated with either EtOH (control) or 4OHT (Zbtb11 KO) for 48 h. Plot shows log2-transformed fold-change (log FC) against transcript abundance (mean counts per million, log CPM) for all genes expressed in ESCs. Differentially expressed (DE) genes (FDR < 0.05) are shown in red. b Zbtb11 expression rescue experiment. Zbtb11lox/lox Rosa26ERt2-Cre cells were transfected with a plasmid expressing either FLAG-Zbtb11-IRES-GFP or GFP alone as control, and treated with either EtOH (Control) or 4OHT (Zbtb11 KO). GFP-positive cells were FACS-sorted 48 h post treatment. Left panel—immunoblot of whole-cell lysates showing the expression of Zbtb11 protein was specifically restored. Right panel—qRT-PCR measuring the expression of a panel of genes downregulated in Zbtb11 KO cells. Barplot shows mean and standard deviation of n = 5 independent experiments. Source data are provided as a Source Data file. c Conservation of Zbtb11 binding at promoters of Zbtb11-dependent genes. Plots show strength of Zbtb11 binding at conserved target promoters in mESC, thymocytes and HEK293 cells (Fig. 1f). Binding sites at promoters of genes DE in Zbtb11 KO cells are marked in red—note these are the strongest sites in the dataset and very well conserved across cell types and species. r = Pearson’s correlation coefficient; p = p value for two-sided correlation test based on Pearson’s product–moment correlation coefficient. d DAVID functional annotation analysis of the genes differentially expressed in Zbtb11 KO indicates one significant cluster of annotation terms (FDR < 0.05), which is related to mitochondria. Significance (−log10 FDR) is plotted against enrichment of the annotation terms associated with the genes. e Immunoblots for two mitochondrial proteins (Mrpl48 and Ndufc2) for which transcripts were found to be downregulated in Zbtb11 KO cells. Lamin B was used as a loading control.