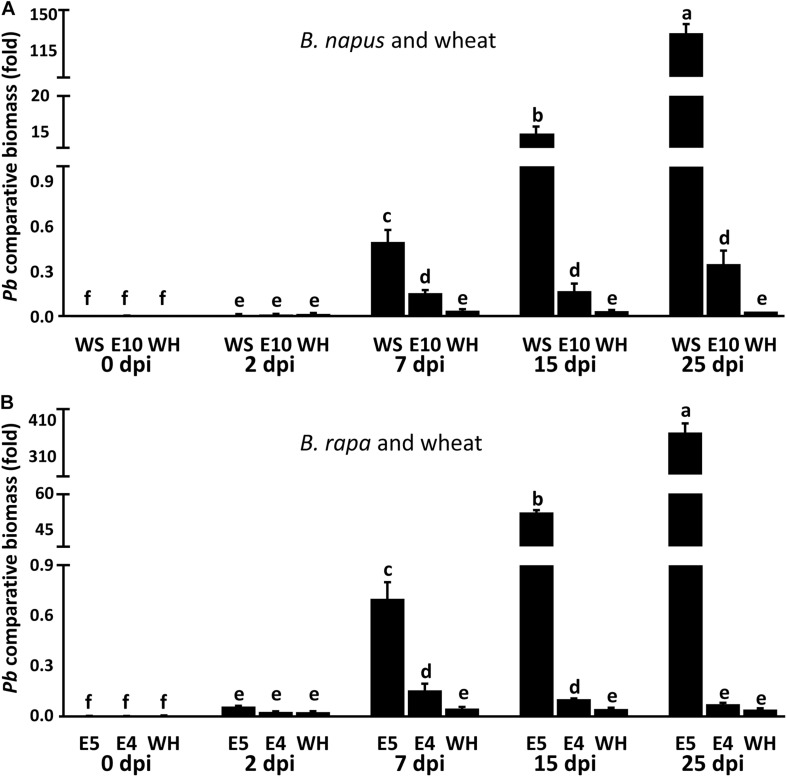
FIGURE 7.
Determination of P. brassicae biomass in susceptible and resistant hosts and non-hosts by qPCR. Roots of susceptible hosts Westar and ECD5, resistant hosts ECD10 and ECD4 and non-host crop wheat were collected at 0, 2, 7, 15, and 25 dpi with three repeats, respectively. The total DNA of each root sample was subject to qPCR analysis using specific primers. The primers designed by the sequence of ITS1 and 5.8S rDNA were used for the amplification of P. brassicae. The actin genes of B. napus, B. rapa, and wheat were used as the internal controls. (A) The biomass of P. brassicae in susceptible and resistant hosts of B. napus and non-host wheat. (B) The biomass of P. brassicae in susceptible and resistant hosts of B. rapa and non-host wheat. The different letters denote statistically significant difference according to one-way ANOVA followed by Tukey’s test (p < 0.05). All the primers used here were listed in Supplementary Table S2. WS, Westar; E10, ECD10; WH, wheat; E5, ECD5; E4, ECD4; Pb, P. brassicae.