Fig. 2.
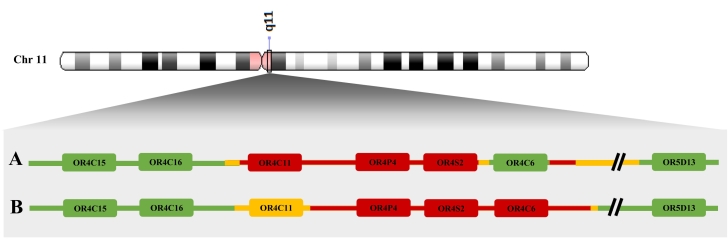
Detected copy number variable region in current and initial 11q11 copy number variant loss study.
Comparison of the 11q11 deletion discovered in (A) our study by multiplex amplicon quantification and (B) the study of Jarick et al. (2011) by the Affymetrix Genome-Wide Human CNV Array 6.0. Red coloring represents the minimal deleted region while yellow coloring signifies the maximal deleted region. Green coloring indicates genes that are not positioned in the deletion. Differences in size and chromosomal location are observed. In the current study, ± 80 kb spanning chr11:55,368,225-55,448,559 was identified as minimal region and a ± 147 kb deletion spanning chr11:55,356,922-55,504,384 as maximal region. An internally retained segment was present with ±1.3 kb as minimal size spanning chr11:55,432,491-55,433,765 and ± 16 kb as maximal size spanning chr11:55,419,331-55,435,229. In contrast, Jarick et al. (2011) identified a minimum size deletion of ±80 kb spanning chr11:55,374,020-55,453,589 and a maximum size deletion of ±97 kb spanning chr11: 55,363,328-55,460,696. Both copy number variable regions cover the olfactory receptor genes OR4P4 and OR4S2; inconsistencies are perceived for genes OR4C11 and OR4C6.
HgLiftOver was applied to convert the deleted region of the initial 11q11 CNV loss study from genome build GRCh36/hg18 to GRCh37/hg19.