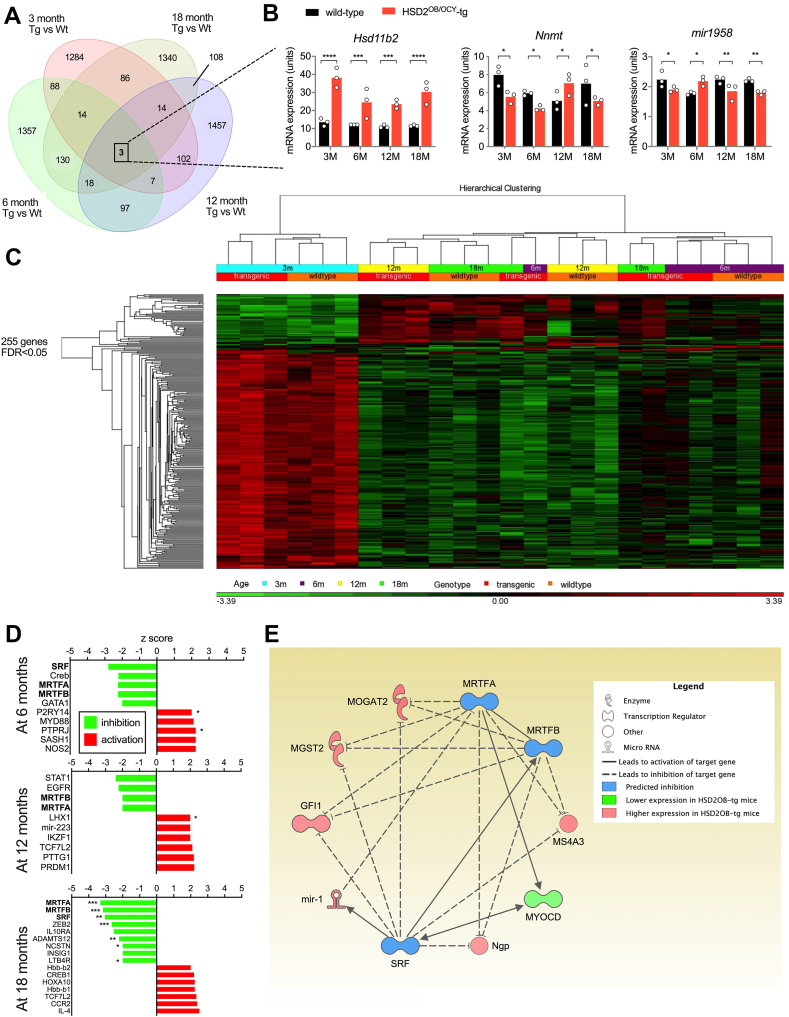
Figure 6.
Skeletal gene expression analysis from wild-type and HSD2OB/OCY-tg mice during aging, and identification of an SRF-MRTFA-MRTFB gene network in aged HSD2OB/OCY-tg mice. (a) Venn diagram of differentially expressed genes in bones from female wild-type and HSD2OB/OCY-tg mice, at unadjusted p < 0.05, n = 3 of each genotype at each timepoint. The number of shared differentially expressed genes at each timepoint (3, 6, 12, 18 months) is shown. (b) Skeletal gene expression levels of Hsd11b2, Nnmt and mir-1958 at 3, 6, 12 and 18 months of age. (c) Hierarchical clustering of 255 differentially expressed genes (false discovery rate <0.05) in female wild-type and HSD2OB/OCY-tg mice at 3, 6, 12 and 18 months of age. Red indicates upregulated and green indicates downregulated (scale is shown below the chart). (d) Z-scores for upstream regulators of gene expression in the bones of HSD2OB/OCY-tg mice, relative to wild-type mice, at 6, 12 and 18 months of age. (e) Putative SRF-MRTFA-MRTFB network at 6–18 months of age in HSD2OB/OCY-tg mice. See legend for explanation of symbols. In all cases, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 and ∗∗∗∗p < 0.0001 for comparisons.