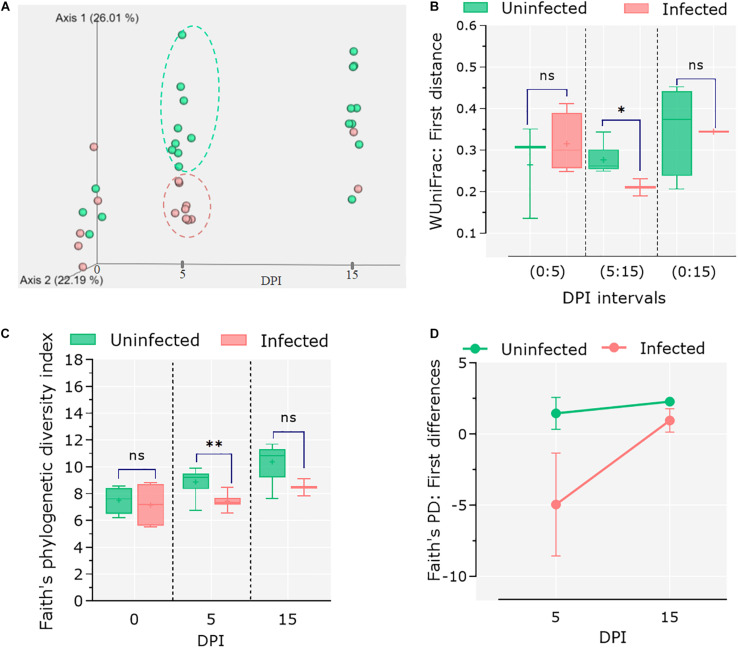
FIGURE 2.
Differential microbial diversity in goat kid microbiota due to Cryptosporidium parvum infection. (A) Principal coordinate analysis (PCoA) of weighted UniFrac distances. We chose to represent the duration of infection on the third principal axis (0, 5, and 15 dpi). Each dot represents an individual microbiome (green = uninfected, red = infected); ellipsoids were arbitrarily drawn to highlight the difference between groups at 5 dpi. (B) Pairwise weighted UniFrac distance (degree of dissimilarity between an individual’s microbiota structure at two separate time points) indicates the distance (dissimilarity) between paired samples from uninfected and infected animals in relation to infection time (dpi). We tested whether these differences were significant at three time intervals (dpi): 0–5, 5–15, and 0–15. (C) Comparison of alpha diversity between uninfected and infected goat kids as measured by the “Faith” phylogenetic index (species richness) at genus level at 0, 5, and 15 dpi. (D) Pairwise difference comparisons addressing the changes in Faith phylogenetic index between paired samples in relation to infection time (i.e., species richness in each individual at 5 dpi in relation to itself at 0 dpi and microbiota differences as compared between infected and uninfected animals). Error bars represent standard deviation. Statistical significance was determined by Kruskal–Wallis test (n.s., ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001).