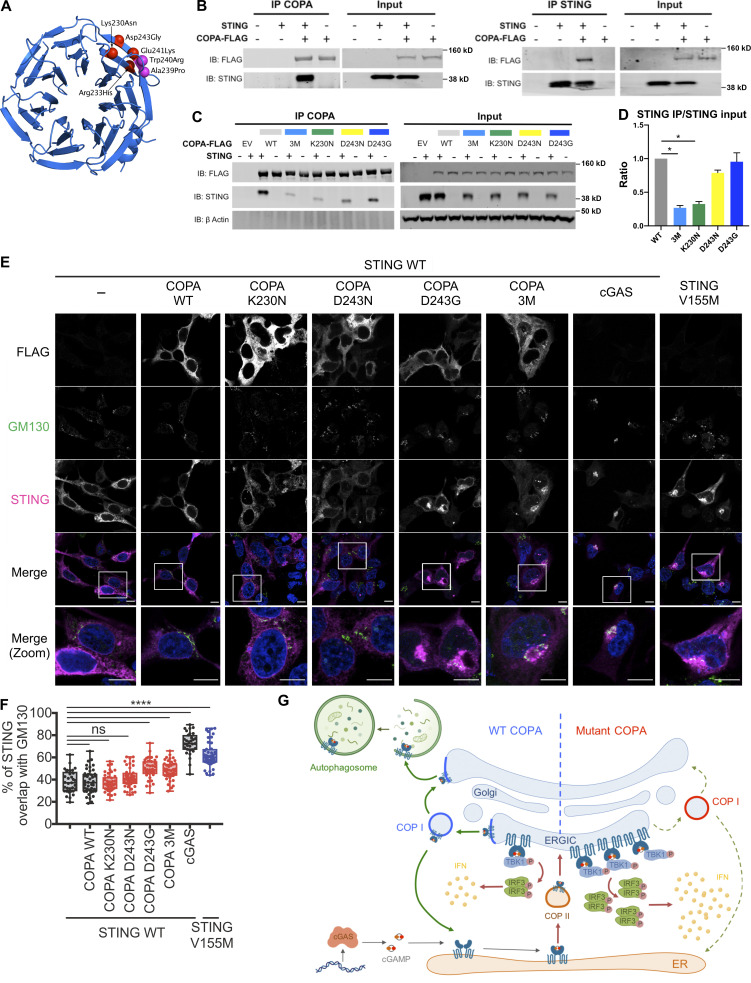
Figure 5.
Interaction of COPA and STING and STING localization. (A) Location of pathogenic missense mutations within the structure of the COPA WD-repeat domain (PDB ID: 6PBG). The four mutations studied in this report are shown in red, while two further previously identified mutations are indicated in magenta. Location of the pathogenic missense mutations within the structure of the COPI coat leaf is given in Fig. S3 B. (B) Western blot analysis of FLAG and STING in proteins IP with an antibody against FLAG (IP COPA, left) or STING (right), and in whole-cell lysates (input) of HEK293T cells cotransfected with EV (−) or WT STING and WT COPA plasmids. Data are representative of three independent experiments. (C) Western blot analysis of FLAG, STING, and β-actin in proteins IP with an antibody against FLAG (IP COPA) and whole-cell lysates (input) of HEK293T cells cotransfected with EV (EV or −) or WT STING and WT COPA, or with individual mutant COPA plasmids (K230N, D243N, D243G) or with a plasmid carrying three substitutions (3M: K230N/R233H/D243N). Data are representative of three independent experiments. (D) Quantification of STING protein levels co-immunoprecipitated (STING IP) with COPA-FLAG compared with the signal recorded in the input, as observed in C. Mean ± SEM of three independent experiments were statistically analyzed using Kruskal–Wallis test (*, P < 0.05). (E) Representative images of STING and COPA localization in HEK293FT cells transfected with WT, mutant COPA (3M carrying three substitutions: K230N/R233H/D243N), and STING plasmids or corresponding control plasmids (−), and transfected with cGAS or control plasmid the next day. Cells were fixed and stained for nuclear DNA (DAPI; blue), COPA (FLAG; gray), Golgi (GM130; green), and STING (magenta). “Merge” row shows an overlay of DAPI, GM130, and STING signals, GM130 and STING costaining being represented in white. “Merge (zoom)” depicts an enlargement of the square above. Images are representative of three independent experiments. Scale bar, 10 µm. (F) Quantification of the ratio of STING signal localized to the GM130/Golgi compartment over total STING signal in images as in A, for at least 10 cells per condition per experiment (one-way ANOVA with Dunnett’s correction: ****, P < 0.0001). ns, not significant. (G) Left: DNA from pathogens and damaged cells induces the production of cGAMP by cGAS. Upon binding of cGAMP, STING translocates, in a COPII-dependent process, to the ERGIC and Golgi, where it triggers TBK1 and IRF3 phosphorylation and subsequent IFN production. We hypothesize that COPA, within the COPI complex, plays a role in STING trafficking after signaling, taking STING back to the ER, or on to endolysosomes/the autophagy pathway (Gonugunta et al., 2017), and thereby leading to resolution of IFN signaling. Our data indicate that STING may be a cargo of COPI through COPA. Right: When COPA is mutated in the cargo-binding WD40 domain, STING can no longer be sorted into COPI vesicles, and so is retained in the ERGIC/Golgi in an active state, leading to continued IFN production. COP, coatomer protein; IB, immunoblot.