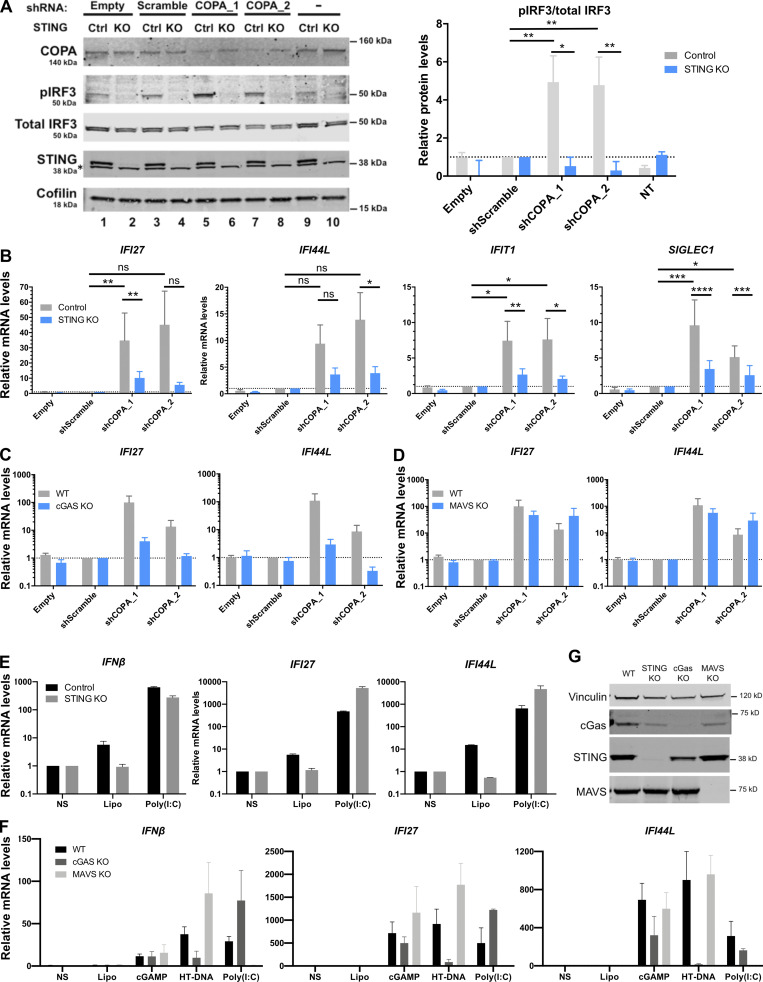
Figure S2.
Knockdown of COPA induces STING-dependent, MAVS-independent type I IFN signaling in THP-1 cells. (A) Left: Western blot analysis of IRF3 phosphorylated at Ser396, total IRF3, COPA, STING, and Cofilin in whole-cell lysates of control or STING KO THP-1 cells transduced with an EV, a scrambled shRNA, or two different shRNAs targeting COPA (shCOPA_1 and shCOPA_2, respectively lanes 5 and 6, and lanes 7 and 8), or nontransduced (NT). Data are representative of three independent experiments. Asterisk indicates unspecific band. Right: Quantification of phosphorylated IRF3 (at position Ser396) relative to total IRF3 expressed as fold over scrambled shRNA relative signal, statistically analyzed in one-way ANOVA (shScramble vs. shCOPA for control THP-1 cells; **, P = 0.0026 for shCOPA_1 and **, P = 0.004 for shCOPA_2) or two-way ANOVA (control vs. STING KO THP-1 cells for each shCOPA; *, P = 0.0104 for shCOPA_1 and **, P = 0.0094 for shCOPA_2) with Sidak’s multiple comparisons test (mean ± SEM). (B) mRNA expression analysis assessed by RT-qPCR of four out of six tested ISGS (IFI27, IFI44L, IFIT1, and SIGLEC1; see other results in Fig. 4 D) in control or STING KO THP-1 cells transduced with an EV, a scrambled shRNA, or shCOPA_1 and shCOPA_2. Mean ± SEM of three independent experiments were statistically analyzed in one-way ANOVA (shScramble vs. shCOPA for control THP-1 cells) or two-way ANOVA (control vs. STING KO THP-1 cells for each shCOPA) with Sidak’s multiple comparisons test (*, P <0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, not significant). (C and D) mRNA expression analysis assessed by RT-qPCR of two ISGs (IFI27, IFI44L; D) in WT or THP-1 cells null for cGAS or MAVS transduced with an EV, a scrambled shRNA, or shCOPA_1 and shCOPA_2. Mean ± SEM of three independent experiments were statistically analyzed in one-way ANOVA (shScramble vs. shCOPA for WT THP-1 cells) or two-way ANOVA (WT vs. MAVS KO THP-1 cells for each shCOPA) with Sidak’s multiple comparisons test (ns, nonsignificant). (E and F) mRNA expression analysis assessed by RT-qPCR of IFNβ and two ISGS (IFI27, IFI44L) in control or THP-1 cells null for STING, cGAS, or MAVS and nonstimulated (NS) or stimulated with 1 µg/ml poly(I:C), 1 µg/ml 2′3′cGAMP, or 0.25 µg/ml HT-DNA for 24 h combined with Lipofectamine (Lipo). Mean ± SEM of two (poly[I:C]) to three (cGAMP, HT-DNA) independent experiments. (G) Western Blot analysis of cGAS, STING, and MAVS expression in THP-1 WT, STING, cGAS, or MAVS KO, and loading control Vinculin. Representative of two independent experiments. Ctrl, control.