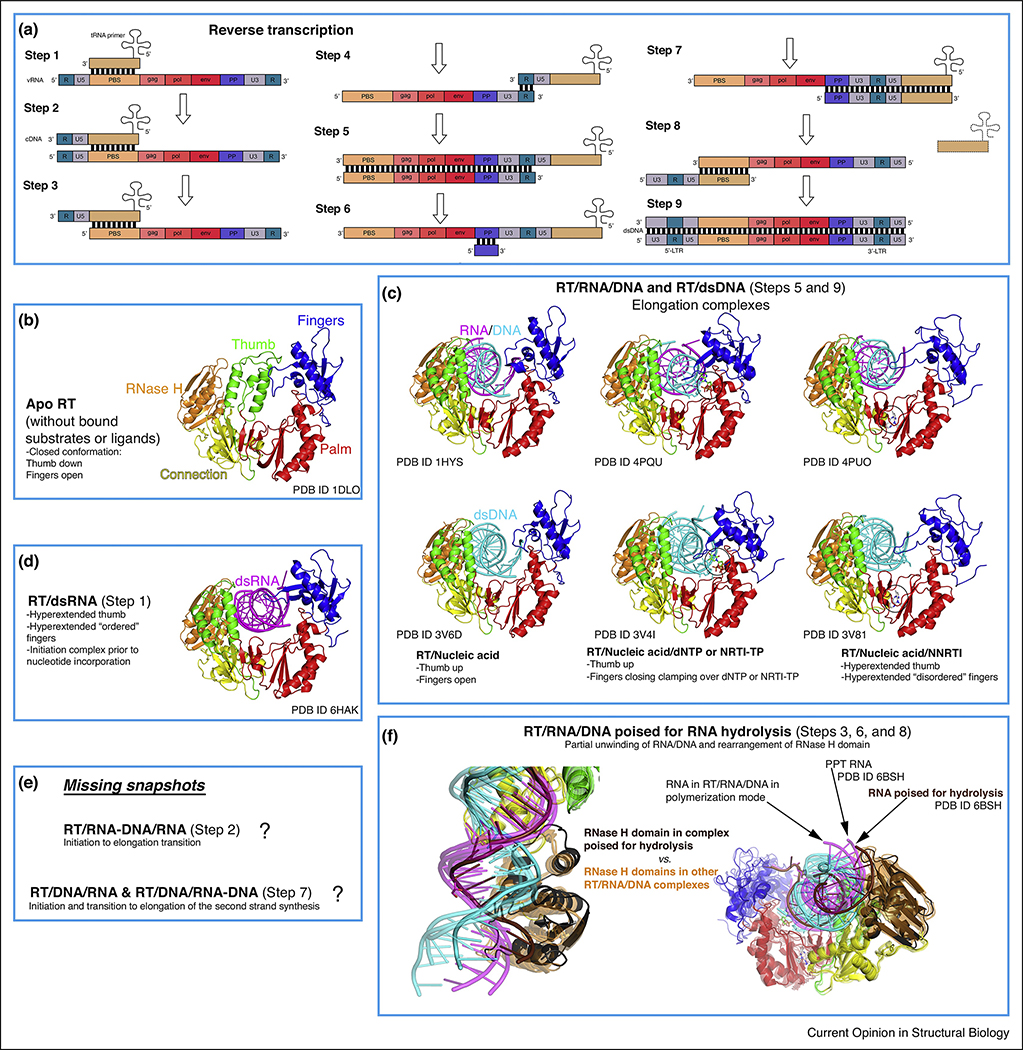
Figure 2. Conformational states of RT during reverse transcription.
(a) Schematic representation of retroviral reverse transcription, displaying the genomic and host nucleic acids (modified from https://en.wikipedia.org/wiki/Reversetranscriptase#/media/File:Reversetranscription.svg, Creative Commons Attribution License CC BY 3.0 license). In brief, the different steps correspond to: 1- Initiation of first strand synthesis; 2- Initiation to elongation transition (RNA/DNA hybrid primer); 3- Hydrolysis of the 5’ region of the viral RNA (vRNA); 4- tRNA jump to the 3’ region of the vRNA; 5- Elongation (dsDNA); 6- vRNA degradation (except polypurine tract, PPT); 7- Initiation of the second strand synthesis; 8- PPT hydrolysis and second jump (tRNA exit); 9- Second strand elongation and addition of the long terminal repeats (LTRs). (b-f) The rest of the boxes (except from the one indicating the challenges remaining, that is, uncharacterized conformational states) contain all the described conformations of RT complexed with nucleic acids (and apo form) during reverse transcription, with color coding and annotations included for clarity. The notion of ‘ordered’ versus ‘disordered’ hyperextended fingers subdomain (boxes C and D) is based on B-factors (lower versus higher) and number of interactions with the nucleic acid strand (more versus less).