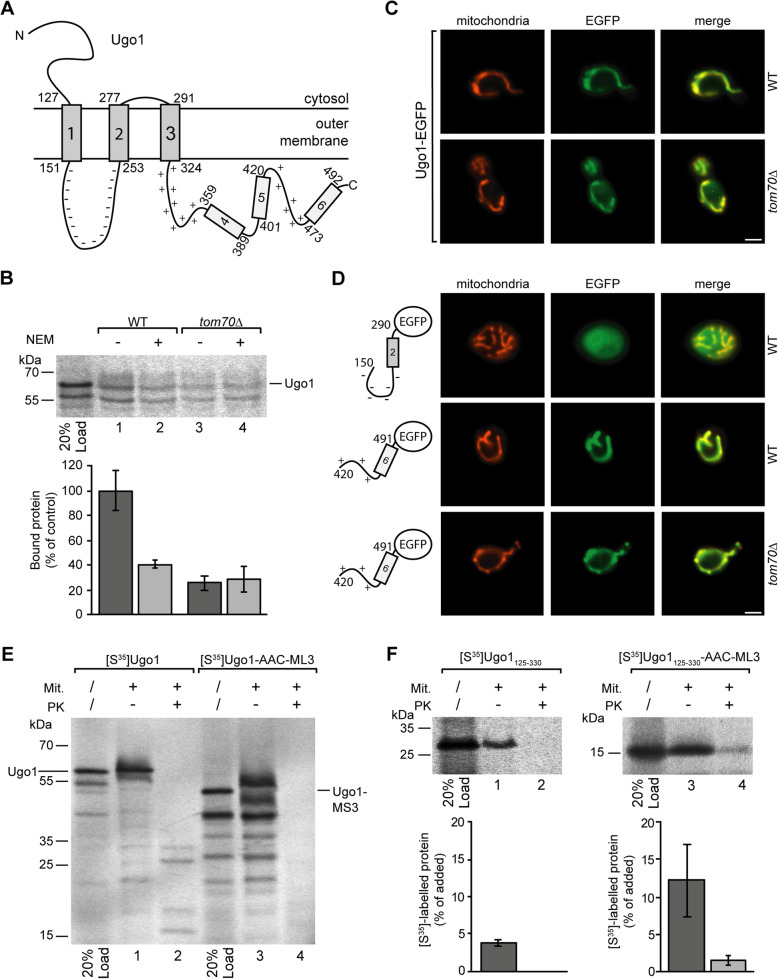
Fig. 6.
Mitochondrial targeting of Ugo1. a Topology of Ugo1 (as described by Hoppins et al., 2009 [60]). b Binding of [35S]-labeled Ugo1 to mitochondria isolated from yeast wild type cells (WT) or from a tom70Δ deletion strain. The mitochondria of samples 2 and 4 were pretreated with 2 mM NEM as described in the legend of Fig. 3. The average amount of Ugo1 bound to wild type mitochondria in the absence of NEM was set to 100% (control), SD, n = 4. c Fluorescence microscopy of EGFP-labeled Ugo1 expressed in yeast wild type cells (upper panel) and in cells of a tom70Δ deletion strain (lower panel). Mitochondria are labeled by MitoTracker Orange; bar 2 μm. d Images of yeast cells expressing different segments of Ugo1 labeled by a EGFP moiety expressed in wild type and in tom70Δ cells, respectively. e Incubation of isolated mitochondria with radiolabeled Ugo1 and a modified version of Ugo1 containing a substitution of residues 151–253 against the matrix loop of AAC module III (ML3, AAC residues 238–277). The mitochondria of samples 2 and 4 were subsequently treated with proteinase K (PK). Following SDS-PAGE, the radiolabeled polypeptides were visualized using the phosphorimager. f A [35S]-labeled fragment of Ugo1 (residues 125–330) and a modified version of this fragment (Ugo1125-330-AAC-ML3) containing the substitution of the negatively charged loop against the matrix loop of AAC module III were incubated with mitochondria as in e. The phosphorimager was used for quantification, and relative amounts of added protein are shown, SD, n ≥ 9