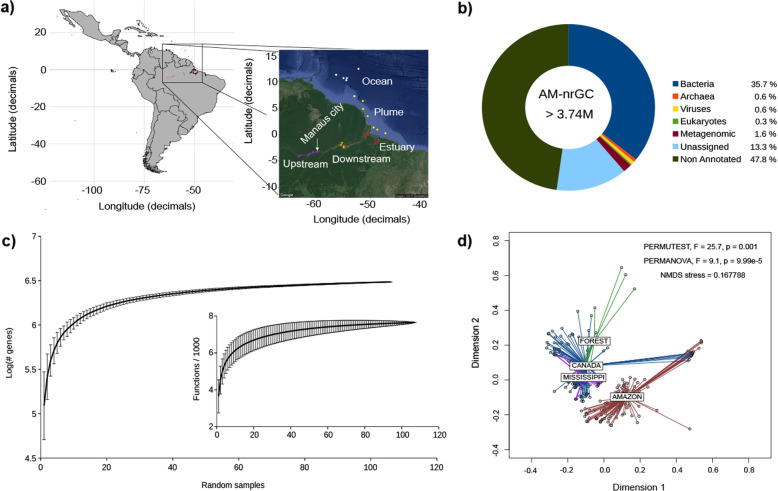
Fig. 1.
The Amazon River basin Microbial Non-Redundant Gene Catalogue (AMnrGC). a Distribution of the 106 metagenomes used in this work over the five sections of the Amazon River: Upstream (purple dots), Downstream (orange dots), Estuary (red dots), Plume (yellow dots) and coastal Ocean (white dots). b Taxonomic classification of the ~ 3.7 million genes in the AMnrGC. “Unassigned” genes were not assigned taxonomy, but they were functionally assigned, differently from “non-annotated” genes, which do not have any ortholog. Those genes displaying orthology to poorly characterized genes found in metagenomes were referred to as “Metagenomic”. c Rarefaction curves of non-redundant genes and PFAM families (internal plot). Note both point towards saturation. d NMDS comparing the Amazon river microbiome with other microbiomes based on information content [k-mer composition]. Amazon River (AMAZON), Amazon forest soil (FOREST), Canada watersheds (CANADA) and Mississippi River (MISSISSIPPI)