Extended Data Figure 8 (related to Figure 3). Depletion of CEP192 in RPE-1 cells recapitulates the synthetic lethal mitotic phenotypes observed in high-TRIM37 expressing cells.
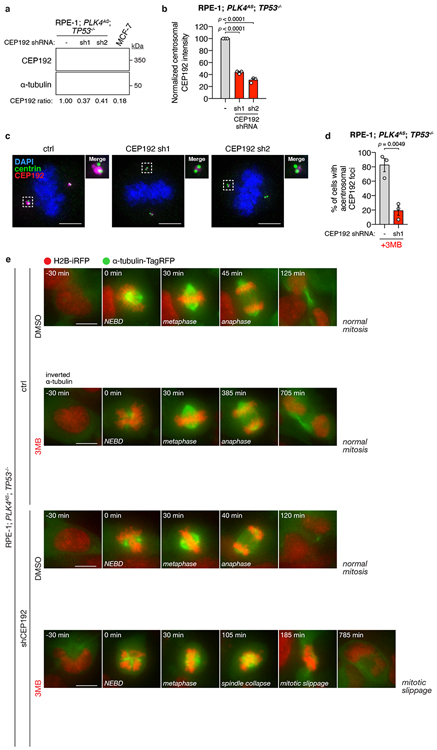
(A) Immunoblot showing the CEP192 levels in indicated control and CEP192-depleted RPE-1; PLK4AS; TP53−/− cells. α-tubulin, loading control. For gel source data, see Supplementary Figure 1.
(B) Quantification of mitotic centrosomal CEP192 signal in the same cells as described in (A). n = 3, biological replicates, each comprising >30 cells. P values, unpaired two-tailed t-test. Mean ± s.e.m.
(C) Representative images of centrosomal CEP192 in the same cells as described in (A). Scale bars, 5 μm.
(D) Quantification of the percentage of 3MB-PP1 treated RPE-1; PLK4AS; TP53−/− cells with acentrosomal mitotic CEP192 PCM foci. n = 3, biological replicates, each comprising >30 cells. P values, unpaired two-tailed t-test. Mean ± s.e.m.
(E) Representative time-lapse images of mitotic progression in DMSO or 3MB-treated control and CEP192-depleted RPE-1; PLK4AS; TP53−/− cells. Cells are labelled with H2B-iRFP and tagRFP-tubulin. n = 3, biological replicates.
