Abstract
Large ovarian follicles are primary characteristics of oviparous species. The development of such follicles is crucially governed by strict intrinsic complex regulation. Many aspects of the genetic basis of this regulation remain obscure. To identify the dominant genes controlling follicular development in the chicken, growing follicles (400–1,600 μm in diameter) were selected for RNA sequencing and bioinformatics analysis. Comparing the 400-μm follicles with 800-μm follicles identified a total of 3,627 differentially expressed genes (1,792 upregulated and 1,835 downregulated genes). Comparing the 400-μm follicles with 1,600-μm follicles revealed 9,650 differentially expressed genes (including 4,848 upregulated and 4,802 downregulated genes). Comparing 800-μm with 1,600-μm follicles revealed a total of 6,779 differentially expressed genes (3,427 upregulated and 3,352 downregulated genes). Transcriptome analysis revealed that genes related to the extracellular matrix–receptor interactions, steroid biosynthesis, cell adhesion, and phagosomes displayed remarkable differential expressions. Relative to 400-μm follicles, collagen content, production of steroid hormones, cell adhesion, and phagocytic factors were significantly increased in the 1,600-μm follicles. This study identifies the dominant genes involved in the promotion of follicular development in oviparous vertebrates and represents the extraordinary gene regulation pattern related to development of the growing follicles in poultry.
Key words: growing follicle, transcriptome, bioinformatics analysis, chicken
Introduction
The process of ovarian follicle maturation in oviparous species requires the nutrients that are required for the early development of the embryo to accumulate in the cytoplasm before egg laying. In poultry breeds, high-efficiency follicular development implies huge economic output for the egg industry. Birds have a unique follicular development process with large amount of yolk in the oocyte cytoplasm (Johnson, 2015, Liu et al., 2018). During follicular development, a series of key events such as gene transcription and protein expression occur in series and are governed by specific gene expression, which is an intrinsic factor regulating follicular recruitment, selection, and apoptosis of follicular cells (Beg et al., 2002).
After activation, the primordial follicle develops into a primary follicle, a process that includes associated morphological changes in which the granulosa cells change from flat to cubic form. Primary follicles are mainly located in the ovarian cortex near the medulla. During the developmental process, both the diameter of oocytes and the volume of follicles increase, and the cytoplasm begins to exhibit a fine foamy shape with obvious nucleoli and with a significant increase in the number of granulosa cells in the mammal (Adhikari and Liu 2009). In the ovarian cortex of the peak laying hens, many follicles exist concurrently at different developmental stages. These are generally divided into 2 categories based on their diameters: prehierarchical and hierarchical follicles. The diameter of follicles increases to about 40 mm, and the weight of the follicles can attain to about 16 g at ovulation (Okumura 2017). Growing follicles are usually present in the ovarian cortex and can be isolated by enzymatic digestion and mechanical separation (Zhao et al., 2018). Using these isolated follicles, analysis of the functional differences and regulation of development becomes possible. However, extensive clarification of this process on a genetic level has not yet been achieved.
The development of ovarian follicles is controlled essentially by complex mechanisms involving multiple genes. Extensive, rather than selective, genetic comparisons will be likely required to clarify multiple regulations by hormones, growth factors, and cytokines that are involved in follicular development. Therefore, we adopted an RNA sequencing (RNA-seq) technology to evaluate the predominant genes involved in the development of growing follicles.
With the development of genomic research, transcriptomics is one of the most widely used technologies (Costa et al., 2010) that has been already used in studies of porcine growth (Adapala and Kim, 2017), chicken skeletal muscle development (Li et al., 2012b), screening of sperm motility regulators (Sun et al., 2019), human aging (Xiao et al., 2018), poultry disease screening (Matulova et al., 2012), and general genetic diversity (Li et al., 2012a). In poultry, most transcriptome studies focus on growth traits (Allais et al., 2018) and meat quality (Teng et al., 2019). In hens, transcriptome sequencing revealed the genetic parameters of feed efficiency traits in the laying period and its relationship with egg quality traits in the duck (Zeng et al., 2018).
The RNA-seq method was also used for studies on the reproductive tract of the chicken to delineate poultry fertility (Froman et al., 2006). We considered that this technique would be suitable to help reveal the whole-gene expression profiles of growing follicles and the key pathways for regulation of growing follicle development.
In this study, RNA-seq, bioinformatics analysis, and validating experiments were performed to identify the differentially expressed genes between different follicular developmental stages to reveal the molecular mechanisms involved in the development of growth follicles. The results will elucidate the overall gene expression profiles controlling progression of the growing follicles and provide a comparative observation relating to the developmental changes between avian species and mammals.
Materials and methods
Animal
Hy-Line white laying hens (aged 12∼15 wk) were purchased from a local poultry farm and subsequently raised on the university campus farm with free access to food and water. All animal experiments were performed in accordance with the recommendations in the Animal Care and Use Guidelines and were approved by the Animal Care and Use Committee on the Ethics of Animal Experiments of Zhejiang University.
Separation of Growing Follicles and Morphological Observations
Ovaries (n = 9) were obtained from 280-day-old Hy-Line hens under sterile conditions, and extra tissues were removed using fine tweezers and scalpels. The intact ovaries were taken out and washed 3 times with PBS. Then, ovaries were washed in dulbecco's modified eagle medium (Hyclone, Logan, UT) supplemented with 200 IU/mL of penicillin and 200 μg/mL of streptomycin. In brief, the ovary was cut into 6 or 7 pieces, and individual follicles (400–1,600 μm in diameter) were mechanically isolated using insulin gauge needles in dissection medium (Leibovitz's L-15 medium [Hyclone]) supplemented with 5% fetal bovine serum (HyClone, Tauranga, New Zealand), 100 IU/mL of penicillin, and 100 μg/mL of streptomycin. The isolated follicles were picked up with the aid of a mouth-operated micropipette under a stereomicroscope, and the number and diameter of isolated follicles were recorded. The follicle size was measured using a micromirror in a stereoscope, and the follicles were divided into 3 groups with a diameter of 400 (±10%), 800 (±5%), and 1,600 (±5%) μm, respectively (Zhao et al., 2018).
The follicles were fixed in 4% neutral paraformaldehyde solution at 4°C for 24 h, dehydrated in graded ethanol, then clarified in xylene, and embedded in paraffin. The embedded samples were then sliced at 5-μm thickness for hematoxylin and eosin staining. Hematoxylin and eosin staining was performed using standard protocols.
Masson Staining
The paraffin section was dewaxed for Lichun red acid magenta solution staining for 30 s and rinsed with 0.2% glacial acetic acid. After treatment with phosphomolybdic acid for 3–5 min, the sections were stained with bright green dye for 10 min, followed with 0.2% glacial acetic acid for 1 min.
Library Preparation for Transcriptome Sequencing
Total RNA was extracted from follicles (5 from every group) using TRIzol (Takara, Shiga, Japan). Total RNA (3 μg) was used as the input material for RNA sample preparation, and mRNA was purified using poly-T oligo-attached magnetic beads. Fragmentation was carried out using divalent cations under elevated temperature in the NEBNext First Strand Synthesis Reaction Buffer (NEB, Ipswich, MA). First-strand cDNA was synthesized using the random hexamer primer and M-MuLV reverse transcriptase (RNase H-). Second-strand cDNA synthesis was subsequently performed using DNA polymerase I and RNase H. Remaining overhangs were converted into blunt ends based on their exonuclease/polymerase activities. After the adenylation of the 3′ ends of the DNA fragments, a NEBNext Adaptor (NEB) with a hairpin loop structure was ligated to prepare for hybridization. To preferentially select cDNA fragments of preferentially 250–300 bp in length, the library fragments were purified using the AMPure XP system (Beckman Coulter, Beverly). Then, 3 μl of USER enzyme (NEB) was used with size-selected, adaptor-ligated cDNA at 37°C for 15 min, followed by 5 min at 95°C before polymerase chain reaction (PCR). PCR was then performed with Phusion High-Fidelity DNA polymerase, universal PCR primers, and Index (X) Primer. Finally, PCR products were purified, and library quality was assessed using the Agilent Bioanalyzer 2100 system (Illumina, San Diego, CA).
Sequencing Data Analysis and Time-Course Analysis
The Q20, Q30, and GC contents of the clean data were calculated. All the downstream analyses were based on clean data with high quality. Reference genome and gene model annotation files were downloaded from the genome website. An index of the reference genome was built using Hisat2 (version 2.0.5, Illumina), and paired-end clean reads were aligned to the reference genome using Hisat2. The Cluster Profiler R package (Illumina) was used to test the statistical enrichment of differential expression genes in Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways (Mao et al., 2005). The Science, Technology, Engineering, and Mathematics was used for short time-course analysis.
Analysis of Different Genes and Enrichment
Differential expression analysis of the 2 conditions/groups (2 biological replicates per condition) was performed using the DESeq2 R package (1.16.1, Illumina). DESeq2 provides statistical routines for determining differential expression in digital gene expression data using a model based on negative binomial distribution. The resulting P-values were adjusted using the Benjamini and Hochberg's approach for controlling the false discovery rate. Genes with an adjusted P-value <0.05 found by DESeq2 were assigned as differentially expressed genes.
Quantitative Real-time PCR Analysis
Quantitative real-time PCR (RT-qPCR) was performed using the SYBR Premix Ex Taq TMKit (Takara) on an ABI 7500HT Real-time PCR detection system (Applied Biosystems, Foster City). The qPCR conditions were as follows: 95°C for 10 min and then 40 cycles of 95°C for 30 s, 64°C for 34 s, and 72°C for 30 s. Comparisons of expression levels were determined by 2−ΔΔCt methods normalized to GAPDH. The sequences of the primers are listed in Supplementary Table 1.
Immunohistochemistry
Immunohistochemistry was performed as per conventional procedures. Endogenous peroxidase blockade was performed using 3% hydrogen peroxide, followed by antigen retrieval in a 10 mmol sodium citrate buffer (pH 6.0) for 30 min. Blocking was carried out for 30 min at room temperature in 5% goat serum (Boster Bioengineering Co., Ltd., Wuhan, China). Tissue sections were incubated overnight at 4°C with the primary antibody against rabbit anti-col4A1 (1:100, BA2174, Boster Biological Technology Co., Wuhan, China) and rabbit anti-col6A1 (1:100, ET1612-91, HuaAn Biotechnology Co., Hangzhou, China). A biotinylated secondary antibody was used and then incubated with horseradish peroxidase–conjugated streptavidin. The sections were then exposed to diaminobenzidine for color development. The sections were counterstained with hematoxylin for 3 min.
Measurement of Progesterone Level
To measure the progesterone level, 400- and 1,600-μm follicles were homogenized in ice-cold 5% PBS, and progesterone levels were determined using an ARCHITECT Progesterone Reagent Kit (Abbott, Lisnamuck, Ireland) as per the manufacturer's protocol.
Western Blot
Follicle lysates were prepared using ice-cold RIPA (P00138, Beyotime, Shanghai, China) with protease inhibitors, followed by centrifugation for 30 min. Protein concentration was determined using a BCA Protein Assay Kit (Jiancheng Bioengineering Institute, Nanjing, China). An equal amount of protein was loaded onto an SDS-PAGE gel separated by electrophoresis and transferred to a polyvinylidene difluoride membrane (Millipore, Bedford). After blocking with 5% goat serum, the blot was probed with the corresponding primary antibody against rabbit anti-CYP11A1 (1:500, ER1906-98), rabbit anti-CYP17A1 (1:500, ET7107-61), rabbit anti-CYP19A1 (1:500, ER1802-38), rabbit anti–E-cadherin (1:500, ET1607-75), rabbit anti-CD68 (1:500, ET1611-53, HuaAn Biotechnology Co., Hangzhou, China), and mouse anti–β-actin (1:1000, ab8226, Abcam, San Francisco, CA). Horseradish peroxidase–conjugated goat anti–rabbit or anti–mouse IgG (sc-2004 or sc-2005, Santa Cruz Biotechnology, Dallas) was exposed using a ChemiScope 3,400 Mini (Clinx, Shanghai, China). The band intensity was quantified using Image J software (National Institutes of Health, Bethesda, MD), and the results were normalized to β-actin.
Statistical Analysis
All experiments were repeated 3 times. Data were analyzed by one-way ANOVA with post hoc Dunnett's test and independent samples t-test using GraphPad Prism7 software (GraphPad Software, San Diego, CA) and presented as mean ± SEM. P < 0.05 was considered to be statistically significant.
Results
Morphology of Growing Follicles
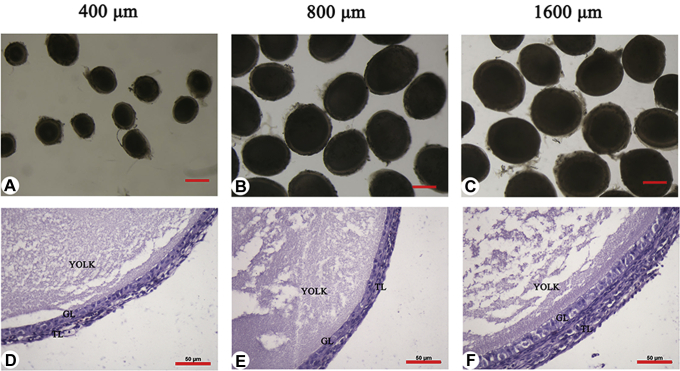
To explore the developmental mechanisms of growing follicles, we isolated 400-, 800-, and 1,600-μm follicles from the hen ovaries. The individual follicles separated by mechanical methods were oval and intact without attached tissues (Figure 1A–C). We observed the morphological features of 400-, 800-, and 1,600-μm follicles after sectioning and hematoxylin and eosin staining. In all 3 categories of follicles, both granulosa and theca layers were clear and intact (Figure 1D–F).
Figure 1.
Morphology of growing follicles in the chicken. (A–C) Three categories of growing follicles. Scale bar: 500 μm. (D–F) H&E staining of the growing follicles. Scale bar: 50 μm. Abbreviations: GL: granulosa layer; TL: theca layer; H&E: hematoxylin and eosin.
Comparison of RNA-seq Data and Reference Genome Sequence
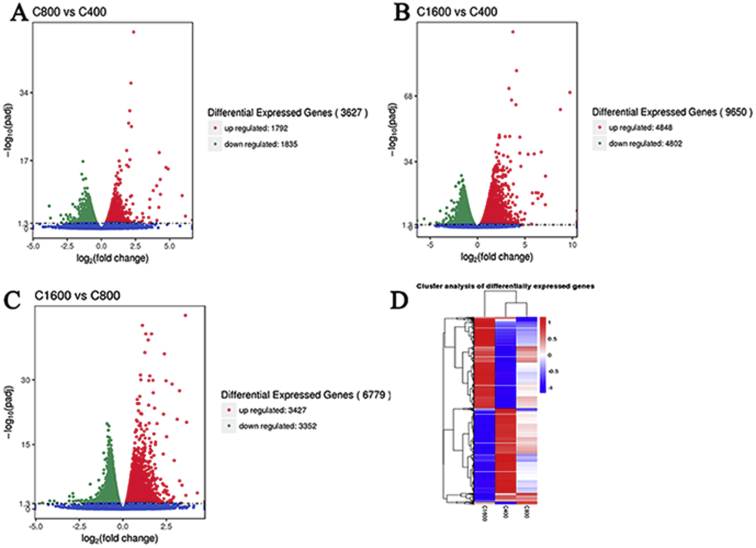
By RNA-seq analysis, 9 cDNA libraries of 400 μm follicles (named C400_1,2,3), 800 μm follicles (named C800_1,2,3), and 1,600 μm follicles (named C1,600_1,2,3) produced 439.1 million original pairs. After quality control, each sample produced a clean read length of approximately 0.48 billion, ranging from 0.423 to 0.587 billion. By comparing clean reads with the reference genome, we obtained mapped reads and mapped data. The unique mapped rates of 3 groups were 87.3, 87.97, and 88.12%, respectively. The ratios of the multiple map of each group accounted for 2.02, 2.14, and 2.08%, respectively (Supplementary Table 2, Supporting Information). We screened for differentially expressed genes between each paired sample set. Compared with 400-μm follicles, there were 1,792 upregulated and 1,835 downregulated genes in the 800-μm follicles and 4,848 upregulated and 4,802 downregulated genes in 1,600-μm follicles. Compared with 800-μm follicles, there were 3,427 upregulated and 3,452 downregulated genes in 1,600-μm follicles (Supplementary Figure 1A–C). The clustering results showed that the gene expression pattern of the 400-μm follicles had a higher similarity to that of the 800-μm than to that of the 1,600-μm follicles. A considerable portion of the gene expression levels of the 400- and 1,600-μm follicles was higher than those of the 800-μm follicles (Supplementary Figure 1D). These results suggested greater changes in the gene expression profiles between follicles of larger differences in diameter.
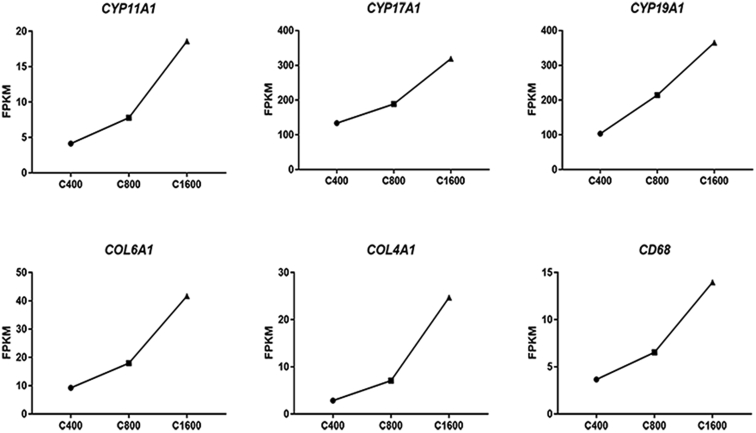
Time-Course Analysis
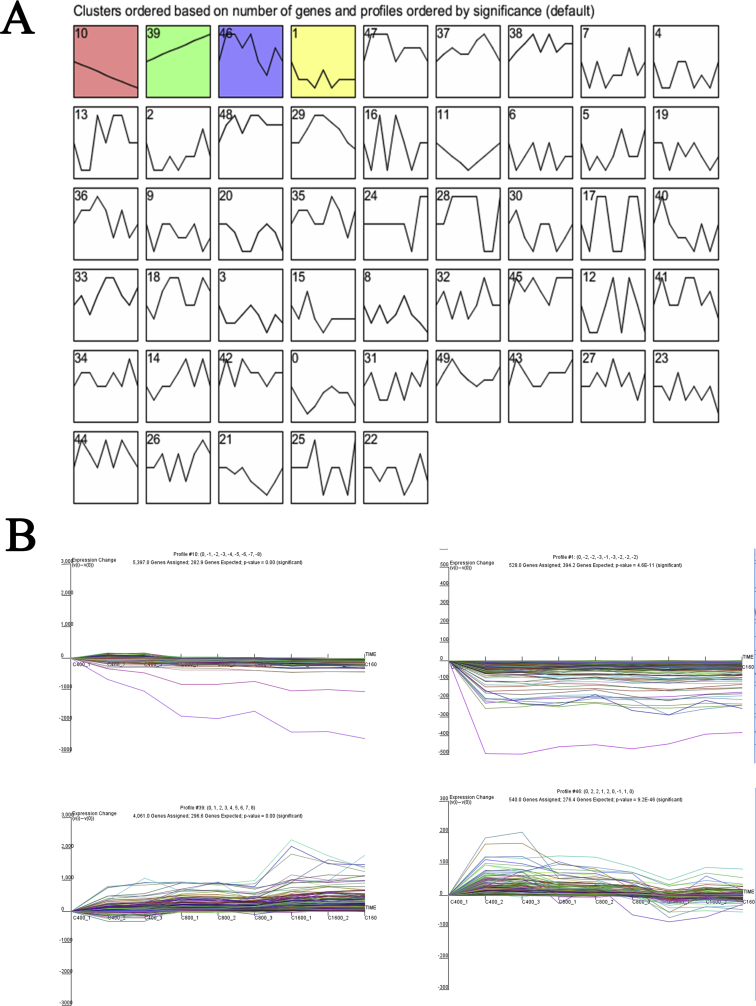
The data set was filtered to contain only 22,056 genes with no missing data. The number in the top left-hand corner of a profile box is the profile ID number. The colored profiles had a statistically significant number of genes assigned (Supplementary Figure 2). Nonwhite profiles of the same color represent profiles grouped into a single cluster. The fragments per kilobase per million of COL4A1, COL6A1, CYP11A1, CYP17A1, CYP19A1, and CD68 was increasing in accordance with the progressive follicular development (Figure 2).
Figure 2.
Time-course analysis. Trends of key gene expression during follicular development by RNA-seq. Abbreviations: FPKM: fragments per kilobase per million; RNA-seq: RNA sequencing.
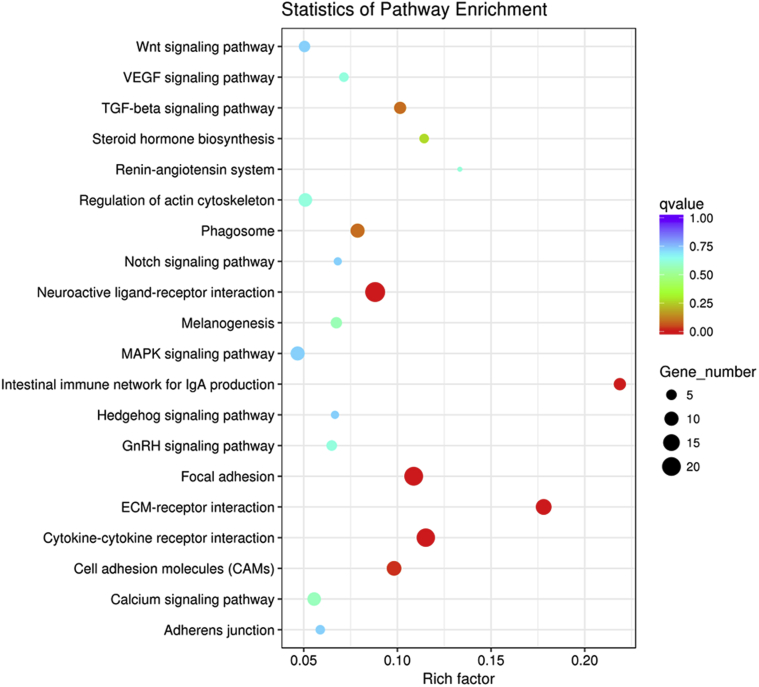
KEGG Analysis of 400- and 1,600-μm Follicles
As per the aforementioned result of the greater difference between the 400- and 1,600-μm follicles than between 400- and the 800-μm follicles, in the subsequent analysis, we only compared the 400-μm follicles with 1,600-μm follicles. Analysis of the pathways enriched by differentially expressed genes in the 400- and 1,600-μm follicles as per the KEGG databases revealed that 78 pathways were enriched. Most notably, 20 of these were KEGG pathways (Figure 3).
Figure 3.
KEGG analysis of differentially expressed genes between 400- and 1,600-μm follicles. The top 20 pathways of 1,600-μm follicles are shown. The size of dot indicates the number of differential genes. The y-axis and x-axis indicates functional pathways and rich factor. Abbreviations: ECM: extracellular matrix; GnRH: gonadotropin-releasing hormone; KEGG: Kyoto Encyclopedia of Genes and Genomes; TGF: transforming growth factor; VEGF: vascular endothelial growth factor.
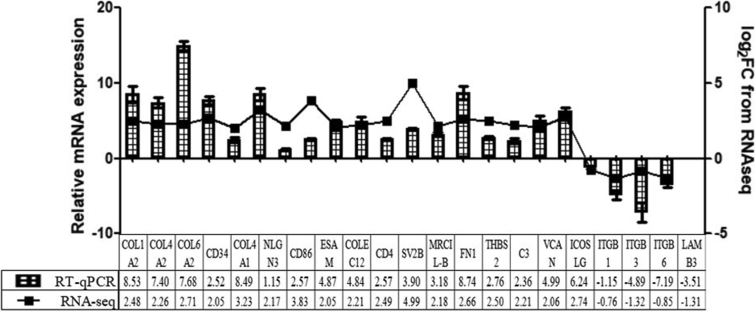
Verification of RNA-seq Data by RT-qPCR
To further evaluate the accuracy of the RNA-seq data, 21 genes were randomly selected and confirmed by RT-qPCR. As shown in Figure 4, the expression trend of the selected genes after RT-qPCR detection was consistent with the RNA-seq analysis, indicating the high accuracy and quality of RNA-seq analysis.
Figure 4.
Validation of RNA-seq differential genes by qPCR. The relative expression levels of different expressed genes of qPCR (strip) were compared with those of RNA-seq (line) data. Abbreviations: RNA-seq: RNA sequencing; qPCR: quantitative polymerase chain reaction.
Key Genes Regulate the Development of Growing Follicles
Among all of the differentially expressed genes analyzed by RNA-seq, extracellular matrix (ECM), cell adhesion, phagocytosis, and steroid synthesis were closely related to the size categories of the growing follicles and associated with developmental stages. The main pathways were those of the ECM and steroid synthesis (Tables 1 and 2). As per the KEGG results, the ECM was the most significantly changed pathway during follicle development. COL1A2, COL6A2, COL4A1, COL4A2, and COL6A1 were all upregulated, whereas ITGB6, ITGA1, LAMB3, ITGB3, and LAMA3 were all downregulated (Table 1). Steroid synthesis was identified as the most significant pathway in the KEGG analysis. CYP19A1, CYP21A2, CYP11A1, HSD17B1, and CYP17A1 all displayed differential expression (Table 2).
Table 1.
Differentially expressed genes involved in extracellular matrix–receptor interaction.
| Gene ID | Gene symbol | log2FC | Description |
|---|---|---|---|
| Upregulated genes | |||
| ENSGALG00000009641 | COL1A2 | 2.4785 | Fibrinogen, alpha/beta/gamma chain, C-terminal globular domain |
| ENSGALG00000011778 | ITGB5 | 2.3801 | Integrin beta subunit, tail |
| ENSGALG00000006859 | SV2B | 4.9945 | Major facilitator superfamily |
| ENSGALG00000006126 | COL6A2 | 2.7083 | von Willebrand factor, type A |
| ENSGALG00000016841 | COL4A1 | 3.2278 | Collagen IV, noncollagenous |
| ENSGALG00000016843 | COL4A2 | 2.2584 | C-type lectin fold |
| ENSGALG00000011200 | THBS2 | 2.5021 | Laminin G domain |
| ENSGALG00000005974 | COL6A1 | 2.2881 | von Willebrand factor, type A |
| ENSGALG00000003578 | fn1 | 2.6556 | Fibronectin, type II, collagen-binding||Fibronectin type III||Fibronectin, type I||Kringle-like fold |
| ENSGALG00000005180 | COL11A1 | 1.913 | Laminin G domain |
| ENSGALG00000008141 | COL4A5 | 0.74003 | Collagen IV, noncollagenous |
| ENSGALG00000008439 | CD36 | 1.1133 | CD36 antigen |
| ENSGALG00000002893 | STC2 | 1.8295 | Stanniocalcin |
| ENSGALG00000016480 | SDC1 | 1.4921 | Neurexin/syndecan/glycophorin C |
| ENSGALG00000002552 | COL3A1 | 2.326 | Collagen Ⅲ |
| ENSGALG00000008141 | COL4A5 | 0.74003 | Collagen IV, noncollagenous |
| ENSGALG00000008266 | COL4A6 | 1.0409 | Collagen triple helix repeat |
| Downregulated genes | |||
| ENSGALG00000011141 | ITGB6 | −0.84832 | von Willebrand factor, type A||Integrin beta subunit, cytoplasmic domain |
| ENSGALG00000014891 | ITGA1 | −0.76116 | von Willebrand factor, type A||Integrin alpha chain||Integrin alpha-2 |
| ENSGALG00000001343 | LAMB3 | −1.3133 | Laminin, N-terminal||Galactose-binding domain-like||EGF-like, laminin |
| ENSGALG00000019761 | CHAD | −0.58267 | Leucine-rich repeat-containing N-terminal||Leucine-rich repeat, typical subtype||Leucine-rich repeat||Cysteine-rich flanking region, C-terminal |
| ENSGALG00000000379 | ITGB3 | −1.3161 | EGF-like domain, extracellular||Plexin-like fold||Integrin beta subunit|| |
| ENSGALG00000015056 | LAMA3 | −0.60567 | Laminin B type IV||Laminin domain II||Laminin G domain||Laminin I||Concanavalin A-like lectin/glucanase domain||EGF-like, laminin |
| ENSGALG00000003147 | TRPC4AP | −0.35023 | Protein of unknown function DUF3689||Armadillo-type fold |
| ENSGALG00000016155 | COL19A1 | −0.76912 | Collagen triple helix repeat||Laminin G domain||Concanavalin A-like lectin/glucanase domain |
| ENSGALG00000013587 | COL2A1 | −1.5671 | Fibrillar collagen, C-terminal||Collagen triple helix repeat||Fibrinogen, alpha/beta/gamma chain, C-terminal globular domain |
| ENSGALG00000027526 | COL9A2 | −1.271 | Collagen triple helix repeat |
Table 2.
Differentially expressed genes involved in steroid hormone biosynthesis.
| Gene ID | Gene symbol | log2FC | Description |
|---|---|---|---|
| Upregulated genes | |||
| ENSGALG00000013294 | CYP19A1 | 1.9383 | Cytochrome P450||Cytochrome P450, E-class, group I||Cytochrome P450, E-class, group IV |
| ENSGALG00000010153 | CYP21A2 | 1.9274 | Cytochrome P450||Cytochrome P450, E-class, group I||Cytochrome P450, E-class, group IV||Cytochrome P450, B-class |
| ENSGALG00000002024 | COMT | 0.48807 | S-adenosyl-L-methionine-dependent methyltransferase||O-methyltransferase, family 3||Catechol O-methyltransferase, eukaryotic |
| ENSGALG00000001417 | CYP11A1 | 2.2882 | Cytochrome P450, mitochondrial||Cytochrome P450||Cytochrome P450, E-class, group IV||Cytochrome P450, E-class, group I||Cytochrome P450, B-class |
| ENSGALG00000007986 | MOCS3 | 0.26973 | Molybdenum cofactor biosynthesis, MoeB||UBA/THIF-type NAD/FAD binding fold||Rhodanese-like domain||MoeZ/MoeB |
| ENSGALG00000027429 | HSD17B1 | 4.1101 | Glucose/ribitol dehydrogenase||Polyketide synthesis, ketoreductase domain||Short-chain dehydrogenase/reductase SDR||2,3-dihydro-2,3-dihydroxybenzoate dehydrogenase |
| ENSGALG00000008121 | CYP17A1 | 1.3743 | Cytochrome P450||Cytochrome P450, B-class||Cytochrome P450, E-class, group I||Cytochrome P450, E-class, group IV |
| ENSGALG00000011894 | CYP2D6 | 0.77783 | Cytochrome P450||Cytochrome P450, E-class, group I, CYP2D-like||Cytochrome P450, E-class, group I||Cytochrome P450, E-class, group IV |
| ENSGALG00000004501 | CYP1C1 | 2.0336 | Cytochrome P450||Cytochrome P450, B-class||Cytochrome P450, E-class, group IV||Cytochrome P450, E-class, group I |
| ENSGALG00000011812 | SULT1E1 | 0.60556 | P-loop containing nucleoside triphosphate hydrolase||Sulfotransferase domain |
| ENSGALG00000014764 | HSD3B2 | 2.0746 | Polysaccharide biosynthesis protein, CapD-like domain||NmrA-like domain||RmlD-like substrate binding domain|| |
| ENSGALG00000008178 | CYP2AB2 | 4.0112 | Cytochrome P450, E-class, group I||Cytochrome P450, E-class, group IV||Cytochrome P450, B-class||Cytochrome P450 |
| ENSGALG00000013535 | CYP4V2 | 1.9959 | Cytochrome P450||Cytochrome P450, E-class, group IV||Cytochrome P450, E-class, group I||Cytochrome P450, E-class, group II |
| Downregulated genes | |||
| ENSGALG00000004436 | CYP3A4 | −1.1846 | Cytochrome P450||Cytochrome P450, E-class, CYP3A||Cytochrome P450, B-class||Cytochrome P450, E-class, group IV||Cytochrome P450, E-class, group II||Cytochrome P450, E-class, group I |
| ENSGALG00000004449 | cyp3A37 | −1.0404 | Cytochrome P450, B-class||Cytochrome P450, E-class, group IV||Cytochrome P450, E-class, group II||Cytochrome P450, E-class, group I||Cytochrome P450||Cytochrome P450, E-class, CYP3A |
| ENSGALG00000006729 | CYP26A1 | −1.5986 | Cytochrome P450, E-class, group I||Cytochrome P450, E-class, group IV||Cytochrome P450, B-class||Cytochrome P450 |
| ENSGALG00000012070 | CYP24A1 | −0.9489 | Cytochrome P450, E-class, group IV||Cytochrome P450, E-class, group I||Cytochrome P450, E-class, CYP24 A, mitochondrial||Cytochrome P450 |
| ENSGALG00000012791 | TBXAS1 | −1.1106 | Cytochrome P450, B-class||Cytochrome P450, E-class, group II||Cytochrome P450, E-class, group I||Cytochrome P450, E-class, group IV||Cytochrome P450||Cytochrome P450, E-class, CYP3A |
| ENSGALG00000020625 | CYP2J24P | −1.013 | Cytochrome P450, E-class, group I||Cytochrome P450 |
| ENSGALG00000002681 | HSD17B7 | −0.55915 | Short-chain dehydrogenase/reductase SDR||Polyketide synthesis, ketoreductase domain||NAD-dependent epimerase/dehydratase, N-terminal domain||Glucose/ribitol dehydrogenase |
Changes of Collagen Expression During Follicle Development
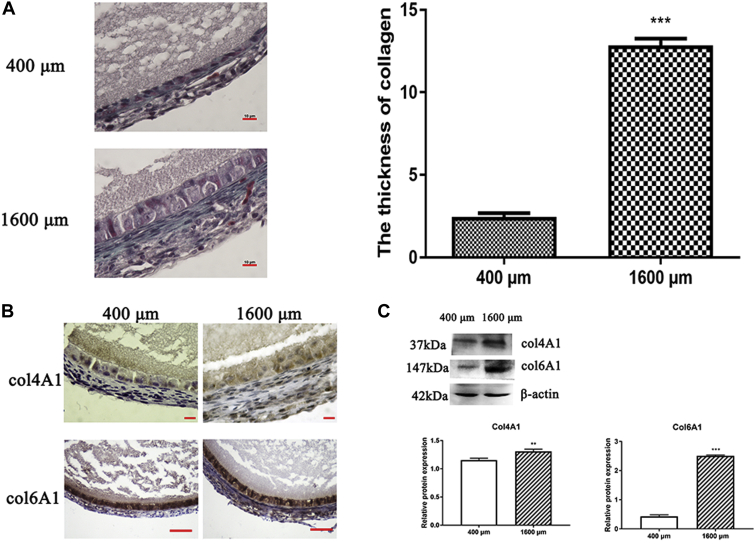
The most important component of the ECM is collagen. The location of collagen was observed by Masson staining. The result showed that collagen in the 400-μm follicles was mainly located in the granulosa layer, whereas in the 1,600-μm follicles, collagen was mainly distributed in the theca layer (Figure 5A). Compared with the 400-μm follicles, the thickness of collagen in the 1,600-μm follicles had significantly increased (Figure 5A).
Figure 5.
Distribution of collagen in 400- and 1,600-μm follicles. (A) Changes of collagen distribution by Masson staining. The thickness of collagen in 400- and 1,600-μm follicles (n = 6). ∗∗∗P < 0.001. (B) Immunohistochemistry staining of Col4A1 in growing follicles. The nuclei of the positive cells show brown color. Scale bars: 10 μm. Immunohistochemistry staining of Col6A1 in growing follicles. The nuclei of the positive cells show brown color. Scale bars: 50 μm. (C) Western blot analysis of col4A1 and col6A1 expression. Values are mean ± SEM (n = 3). ∗∗P < 0.01, ∗∗∗P < 0.001.
As per RNA-seq analysis, type IV and type VI collagen were the predominant types. As shown in Figure 5B, localization of these 2 types of collagen was consistent with the results of Masson staining. The results from Western blot showed that the content of collagens IV and VI in the 1,600-μm follicles was significantly higher than their corresponding levels in the 400-μm follicles (Figure 5C).
Changes of Steroid Synthesis During Follicle Development
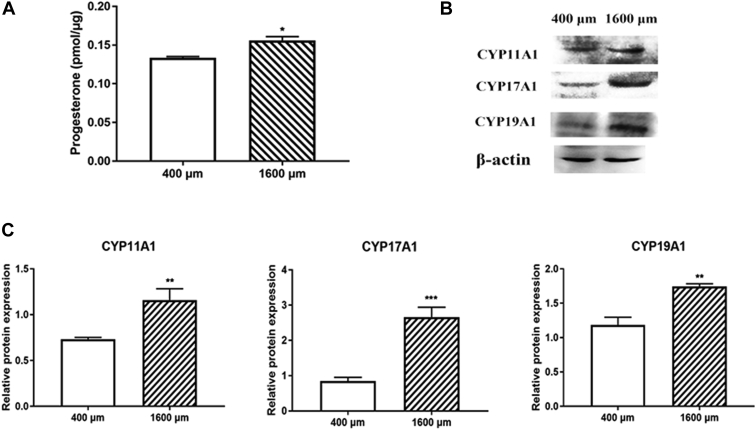
Follicle development is regulated by diverse hormones. KEGG analysis was also enriched in the steroid synthesis pathway. As shown in Figure 6A, the progesterone content of the 1,600-μm follicles was significantly higher than that of the 400-μm follicles (P < 0.05). Western blot analysis revealed that the steroid biosynthesis–related protein including CYP11A1, CYP17A1, and CYP19A1 was higher in the 1,600-μm follicles than that in the 400-μm follicles (Figure 6B, 6C).
Figure 6.
Changes of steroid hormone production in 400- and 1,600-μm follicles. (A) The content of progesterone. ∗P < 0.05. (B and C) Western blot analysis of cyp11A1, cyp17A1, and cyp19A1 expression. Values are mean ± SEM (n = 3). ∗∗P < 0.01, ∗∗∗P < 0.001.
Changes of Cell Adhesion and Phagocytosis During Follicle Development
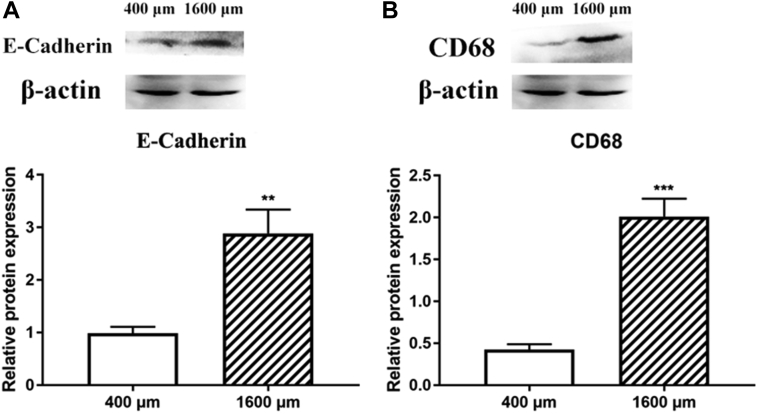
E-cadherin is an important protein for cell adhesion. As shown in Figure 7A, the E-cadherin content in 1,600-μm follicles was higher than that in the 400-μm follicles (P < 0.05). CD68, a marker of phagocytic cells, was also upregulated in the 1,600-μm follicles (Figure 7B).
Figure 7.
Changes of E-cadherin and CD68 in 400- and 1,600-μm follicles. Western blot analysis of E-cadherin (A) and CD68 (B) expression. Values are mean ± SEM (n = 3). ∗∗P < 0.01, ∗∗∗P < 0.001.
Discussion
Analysis of Follicular Development by RNA-seq
Follicle development is a complex physiological process, regulated by diverse genes and endocrine hormones. Illumina's mRNA sequencing technology has enabled and empowered a number of research areas and has led to new discoveries throughout the mRNA field (Wang et al., 2009). In this study, we performed a series of experiments using RNA-seq, bioinformatics analysis, and molecular biochemical techniques to elucidate the developmental mechanisms of growing follicles in laying hen ovaries.
Extracellular Matrices and Development of the Growing Follicles
Extracellular matrices are involved in the regulation of multiple cellular functions in many tissues. They are natural substrates that support cellular processes and are active in regulating cell adhesion, migration, survival, and proliferation (Frantz et al., 2010). Follicle growth and development requires expression of ECMs. The main component of the ECMs is collagen. Previous in vitro studies have shown that mouse ovaries that were cultured on collagen have a significantly higher follicular growth rate than control groups after 10 D of culture (Oktay et al., 2000). Type IV collagen is the main component of the basement membrane and constitutes its skeleton (Niemelä et al., 1990). It not only maintains the integrity of the basement membrane but also plays a key role in its formation. In normal conditions, the basement membrane is stable, dense, and continuous and can prevent macromolecules and cells from passing through. However, tumors can destroy the continuity of this membrane. The increase of collagen type IV is consistent with the degree of hepatic collagen deposition (Niemelä et al., 1990). COL6A1 is a cellular connexin and a major component of the ECM. It is mainly distributed in bone, smooth muscle, and skin tissue. COL6A1 is expressed in a variety of tumors, such as cutaneous fibroids and adolescent angiofibromas (Chen et al., 2013). Collagen VI helps the ECM to form a fibrous network and supports the cellular structure and other cellular components (Owida et al., 2017). Our studies revealed that larger follicles accumulated more collagen, with VI collagen as the predominant type. This result suggests that increased collagen can support the structural integrity of follicles during growth, especially in growing follicles in poultry with increasing diameter and yolk deposition.
Steroid Hormones and Development of the Growing Follicles
Steroid hormone synthetases mainly include the following members: CYP11a1, CYP17a1, CYP19a1, 3β-HSD, and 17β-HSD. Hormones synthesized from steroidogenesis include testosterone, progesterone, and estrogen. In mammals, P450arom was expressed in both granulosa and theca layers of the follicles. However, in poultry, P450arom was only expressed in the theca layers. At the same time, with increasing follicular diameter, the capacity of androstenedione and estradiol synthesis was also enhanced (Shores and Hunter, 1999). Progesterone can regulate follicular development and promote the deposition of yolk material (Abreu et al., 2018). Testosterone and estrogen play pivotal functions in reproduction. Aromatizing enzymes convert testosterone to estrogen. As the volume of follicles increases, the production of progesterone also gradually increases (Jia et al., 2013). This is consistent with our results that an increase in follicle size was accompanied with higher production of sex steroids during development of the growing follicles.
Phagocytosis and Cell Adhesion Molecules With Development of the Growing Follicles
In addition to hormones, cytokines were also involved in the regulation of ovarian development and function. Scattered CD68 cells were also found in human theca and granulosa cell layers (Dahm-Kähler et al., 2009). In this study, we found a higher CD68 content in the larger follicles, indicating a positive relationship between phagocytic activity and follicular development.
Cell adhesion molecules (CAMs) are a type of cell membrane surface glycoprotein. They mediate the interaction between cells and cells or between cells and the ECM. Cell adhesion molecules play an important role in regulating the body shape and maintaining the structural integrity and polarity of adult tissues (Da Silva-Buttkus et al., 2008). E-cadherin is a Ca2+-dependent CAM with the primary function of mediating homologous cell-specific adhesion (Liu, et al. 2017). Expression of E-cadherin was reduced in human breast cancer cells and tissues, making cancer cells more invasive (Tan et al., 2017). E-cadherin regulates intracellular signal transduction pathways such as the inhibition of the WNT/β-catenin pathway (Zhu et al., 2018). Our results showed that E-cadherin protein was elevated in the 1,600-μm follicles in a manner that was consistent with the results of RNA-seq analysis.
The results of the validating experiments by RT-qPCR indicated that there was enrichment of different functional classes in the 3 grades of chicken follicles we studied. This suggests that multiple pathways are involved in the regulation of follicular development. Among these pathways, ECM–receptor interactions, steroid hormone biosynthesis, CAMs, and phagosomes represented the critical and predominant factors involved in promoting development of the growing follicle. Through KEGG analysis, we also found that the gonadotropin-releasing hormone signaling pathway, alpha-linolenic acid metabolism, and linoleic acid metabolism all exhibited decreases during the follicle development process.
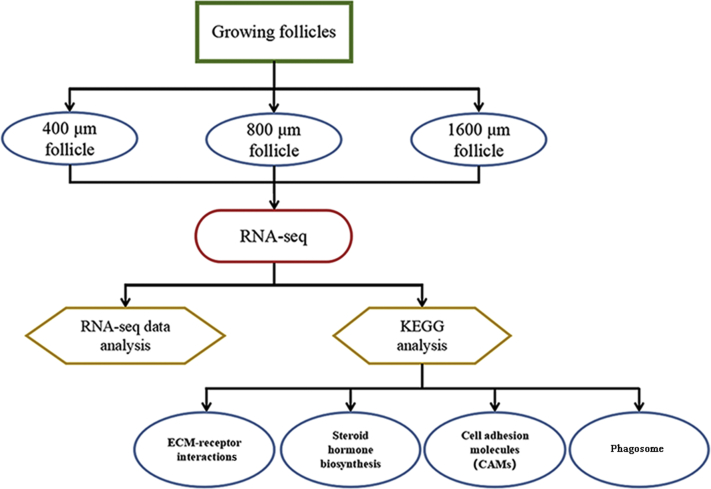
In summary, compared with 400-μm follicles, 9,650 differentially expressed genes were identified in 1,600-μm follicles (4,848 upregulated and 4,802 downregulated). RNA sequencing analysis was further verified by RT-qPCR. Morphological analysis and transmission electron microscopy revealed that rich collagen deposition was observed in 400- and 1,600-μm follicles. Collagen content, steroid hormones, cell adhesion, and phagocytic factors were remarkably increased in 1,600-μm follicles relative to the 400-μm follicles. The aforementioned results indicate that ECM–receptor interactions, steroid hormone biosynthesis, CAMs, and phagosomes play dominant functions in the development of growth follicles in the chicken. These changes meet the requirement of the deposition of large quantity of yolk in the oocytes in oviparous vertebrates, and this serves as an example of an extraordinary gene regulation pattern for the development of growing follicles in poultry (Figure 8).
Figure 8.
Potential regulatory pathways for growing follicles by transcriptome analysis. Abbreviations: CAMs: cell adhesion molecules; KEGG: Kyoto Encyclopedia of Genes and Genomes.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (nos. 31772693, 31972635, and 31472160). The authors are grateful to Animal Science Experimental Teaching Center of Zhejiang University for use of equipment, Weidong Zeng and Dr Jian Li for help in the experiments, and Dr Chris Wood for English improvement in the manuscript.
Conflict of Interest Statement: The authors did not provide a conflict of interest statement.
Footnotes
Supplementary data associated with this article can be found in the online version at http://doi.org/10.1016/j.psj.2019.12.067.
Supplementary data
Supplementary Figure 1.
Volcano map of different expressed genes and gene expression mode in growing follicle. (A–C) Volcano map of differentially expressed genes in the growing follicles. Red dots: up-regulated genes; Green dots: down-regulated genes. (D) Clustering analysis results of gene expression model among 3 categories of growing follicles libraries.
Supplementary Figure 2.
Profiles overview interface. The data were sampled at 3 development time points 400μm,800μm and 1,600μm. The 4 profiles shown were those which had the most genes assigned. All 4 profiles were significant, and the 2 profiles on the left were clustered together. Along the top of each window are statistics on the number of genes assigned to the profile, the number of genes expected, and the enrichment p-value.
References
- Abreu F.M., Geary T.W., da Silva M.A.C., Cruppe L.H., Mussard M.L., Madsen C.A., Martins T., Bridges G.A., Harstine B.R., Day M.L. Role of progesterone concentrations during early follicular development in beef cattle: II. Ovulatory follicle growth and pregnancy rates. Anim. Reprod. Sci. 2018;196:69–76. doi: 10.1016/j.anireprosci.2018.06.011. [DOI] [PubMed] [Google Scholar]
- Adapala N.S., Kim H.K.W. A genome-wide transcriptomic analysis of articular cartilage during normal maturation in pigs. Gene. 2017;27:508–518. doi: 10.1016/j.gene.2017.07.001. [DOI] [PubMed] [Google Scholar]
- Adhikari D., Liu K. Molecular mechanisms underlying the activation of mammalian primordial follicles. Endocr. Rev. 2009;30:438–464. doi: 10.1210/er.2008-0048. [DOI] [PubMed] [Google Scholar]
- Allais S., Hennequet-Antier C., Berri C., Salles L., Demeure O., Le Bihan-Duval E. Mapping of QTL for chicken body weight, carcass composition, and meat quality traits in a slow-growing line. Poult. Sci. 2009;98:1960–1967. doi: 10.3382/ps/pey549. [DOI] [PubMed] [Google Scholar]
- Beg M.A., Bergfelt D.R., Kot K., Ginther O.J. Follicle selection in cattle: dynamics of follicular fluid factors during development of follicle dominance. Biol. Reprod. 2002;66:120–126. doi: 10.1095/biolreprod66.1.120. [DOI] [PubMed] [Google Scholar]
- Chen P., Cescon M., Bonaldo P. Collagen VI in cancer and its biological mechanisms. Trends Mol. Med. 2013;19:410–417. doi: 10.1016/j.molmed.2013.04.001. [DOI] [PubMed] [Google Scholar]
- Costa V., Angelini C., Feis I.D., Ciccodicola A. Uncovering the complexity of transcriptomes with RNA-Seq. J. Biomed. Biotechnol. 2010;2010:853916. doi: 10.1155/2010/853916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Silva-Buttkus P., Jayasooriya G.S., Mora J.M., Mobberley M., Ryder T.A., Baithun M., Stark J., Franks S., Hardy K. Effect of cell shape and packing density on granulosa cell proliferation and formation of multiple layers during early follicle development in the ovary. J. Cell Sci. 2008;121:3890–3900. doi: 10.1242/jcs.036400. [DOI] [PubMed] [Google Scholar]
- Dahm-Kähler P., Ghahremani M., Lind A.K., Sundfeldt K., Brännström M. Monocyte chemotactic protein-1 (MCP-1), its receptor, and macrophages in the perifollicular stroma during the human ovulatory process. Fertil. Steril. 2009;91:231–239. doi: 10.1016/j.fertnstert.2007.07.1330. [DOI] [PubMed] [Google Scholar]
- Frantz C., Stewart K.M., Weaver V.M. The extracellular matrix at a glance. J. Cell Sci. 2010;123:4195–4200. doi: 10.1242/jcs.023820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froman D.P., Kirby J.D., Rhoads D.D. An expressed sequence tag analysis of the chicken reproductive tract transcriptome. Poult. Sci. 2006;85:1438–1441. doi: 10.1093/ps/85.8.1438. [DOI] [PubMed] [Google Scholar]
- Jia Y., Lin J., Mi Y., Zhang C. Prostaglandin E2 and insulin-like growth factor I interact to enhance proliferation of theca externa cells from chicken prehierarchical follicles. Prostag Other Lipid Mediat. 2013;106:91–98. doi: 10.1016/j.prostaglandins.2013.05.007. [DOI] [PubMed] [Google Scholar]
- Johnson A.L. Ovarian follicle selection and granulosa cell differentiation. Poult. Sci. 2015;94:781. doi: 10.3382/ps/peu008. [DOI] [PubMed] [Google Scholar]
- Li Q., Wang N., Du Z., Hu X., Chen L., Fei J., Wang Y., Li N. Gastrocnemius transcriptome analysis reveals domestication induced gene expression changes between wild and domestic chickens. Genomics. 2012;100:314–319. doi: 10.1016/j.ygeno.2012.07.008. [DOI] [PubMed] [Google Scholar]
- Li T., Wang S., Wu R., Zhou X., Zhu D., Zhang Y. Identification of long non-protein coding RNAs in chicken skeletal muscle using next generation sequencing. Genomics. 2012;99:292–298. doi: 10.1016/j.ygeno.2012.02.003. [DOI] [PubMed] [Google Scholar]
- Liu J., Shang D., Xiao Y., Zhong P., Cheng H., Zhou R. Isolation and characterization of string-forming female germline stem cells from ovaries of neonatal mice. J. Biol. Chem. 2017;292:16003–16013. doi: 10.1074/jbc.M117.799403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X.T., Lin X., Mi Y.L., Zeng W.D., Zhang C.Q. Age-related changes of yolk precursor formation in the liver of laying hens. J. Zhejiang Univ-sci B. 2018;19:390–399. doi: 10.1631/jzus.B1700054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao X., Cai T., Olyarchuk J.G., Wei L. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics. 2005;21:3787–3793. doi: 10.1093/bioinformatics/bti430. [DOI] [PubMed] [Google Scholar]
- Matulova M., Rajova J., Vlasatikova L., Volf J., Stepanova H., Havlickova H., Sisak F., Rychlik I. Characterization of chicken spleen transcriptome after infection with Salmonella enterica serovar Enteritidis. Plos One. 2012;7:e48101. doi: 10.1371/journal.pone.0048101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niemelä O., Risteli J., Blake J.E., Risteli L., Compton K.V., Orrego H. Markers of fibrogenesis and basement membrane formation in alcoholic liver disease. Gastroenterology. 1990;98:1612–1619. doi: 10.1016/0016-5085(90)91098-q. [DOI] [PubMed] [Google Scholar]
- Oktay K., Karlikaya G., Akman O., Ojakian G.K., Oktay M. Interaction of extracellular matrix and activin-A in the initiation of follicle growth in the mouse ovary. Biol. Reprod. 2000;63:457–461. doi: 10.1095/biolreprod63.2.457. [DOI] [PubMed] [Google Scholar]
- Okumura H. Avian egg and egg Coat. In: T. Sasanami, editor. Avian Reproduction. 2017. pp. 75–90. [DOI] [PubMed] [Google Scholar]
- Owida H.A., Ruiz T.D.L.H., Dhillon A., Yang Y., Kuiper N.J. Co-culture of chondrons and mesenchymal stromal cells reduces the loss of collagen VI and improves extracellular matrix production. Histochem. Cell Biol. 2017;148:1–14. doi: 10.1007/s00418-017-1602-4. [DOI] [PubMed] [Google Scholar]
- Shores E.M., Hunter M.G. Immunohistochemical localization of steroidogenic enzymes and comparison with hormone production during follicle development in the pig. Reprod. Fertil. Dev. 1999;11:337–344. doi: 10.1071/rd99085. [DOI] [PubMed] [Google Scholar]
- Sun Y., Fu L., Xue F., Li Y., Xu H., Chen J. Digital gene expression profiling and validation study highlight Cyclin F as an important regulator for sperm motility of chickens. Poult. Sci. 2019;0:1–9. doi: 10.3382/ps/pez212. [DOI] [PubMed] [Google Scholar]
- Tan R., Wang L., Song J., Li J., He T. Expression and significance of Twist, estrogen receptor, and E-cadherin in human breast cancer cells and tissues. J. Can Res. Ther. 2017;13:707–714. doi: 10.4103/jcrt.JCRT_1396_16. [DOI] [PubMed] [Google Scholar]
- Teng J., Gao N., Zhang H., Li X., Li J., Zhang H., Zhang X., Zhang Z. Performance of whole genome prediction for growth traits in a crossbred chicken population. Poult. Sci. 2019;98:1968–1975. doi: 10.3382/ps/pey604. [DOI] [PubMed] [Google Scholar]
- Wang Z., Gerstein M., Snyder M. RNA-seq: a revolutionary tool for transcriptomics. Nat. Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao F., Chen X., Yu Q., Ye Y., Liu Y., Yan D., Yang L., Chen G., Lin R., Yang L., Liao X., Zhang W., Zhang W., Tang N.L., Wang X., Zhou J., Cai W., He Y., Kong Q. Transcriptome evidence reveals enhanced autophagy-lysosomal function in centenarians. Genome Res. 2018;28:1601–1610. doi: 10.1101/gr.220780.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng T., Zhang H., Liu J., Chen L., Tian Y., Shen J., Lu L. Genetic parameters of feed efficiency traits and their relationships with egg quality traits in laying period of ducks. Poult. Sci. 2018;97:758–763. doi: 10.3382/ps/pex337. [DOI] [PubMed] [Google Scholar]
- Zhao D., Leghari I.H., Li J., Mi Y., Zhang C. Isolation and culture of chicken growing follicles in 2- and 3-dimensional models. Theriogenology. 2018;111:43–51. doi: 10.1016/j.theriogenology.2018.01.012. [DOI] [PubMed] [Google Scholar]
- Zhu X., Li Y., Zhou R., Wang N., Kang S. Knockdown of E-cadherin expression of endometrial epithelial cells may activate Wnt/β-catenin pathway in vitro. Arch. Gynecol. Obstet. 2018;297:117–123. doi: 10.1007/s00404-017-4560-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.