Abstract
Purpose of this Review:
Human gammaherpesviruses have complex lifecycles that drive their pathogenesis. KSHV and EBV are the etiological agents of multiple cancers worldwide. There is no FDA-approved vaccine for either KSHV or EBV. This review will describe recent progress in understanding EBV and KSHV lifecycles during infection.
Recent findings:
Determining how latency is established, particularly how non-coding RNAs influence latent and lytic infection, is a rapidly growing area of investigation into how gammaherpesviruses successfully persist in the human population. Many factors have been identified as restrictors of reactivation from latency, especially innate immune antagonism. Finally, new host proteins that play a role in lytic replication have been identified.
Summary:
In this review we discuss recent findings over the last 5 years on both host and viral factors that are involved in EBV and KSHV pathogenesis.
Keywords: KSHV, EBV, Latency, Reactivation, Lytic, Tropism
Introduction
Human herpesviruses (HHVs) are separated into three sub-families: α, β, and γ. Two HHVs, Epstein-Barr virus (EBV) and Kaposi’s sarcoma-associated herpesvirus (KSHV), are γ- herpesviruses. Like all herpesviruses, EBV and KSHV, also known as human herpesvirus (HHV)-4 and HHV-8, have a biphasic lifecycle. Both EBV and KSHV rapidly establish latency, wherein a limited subset of viral genes is expressed to modulate the host. Reactivation from latency drives lytic infection to produce new viral progeny that infect new cells or new hosts.
KSHV and EBV can both infect epithelial cells and B cells, while KSHV can infect endothelial cells. Both viruses cause disease in multiple distinct cell lineages. Successfully infecting and establishing latency in distinct cell types requires a large viral investment. Both EBV and KSHV encode multiple proteins and non-coding RNAs to antagonize host defenses and modulate the environment of the cell. Through this host modulation, these viruses contribute to many different disease manifestations. Understanding how these viruses successfully infect cells, establish and maintain latency, and eventually reactivate from latency to trigger lytic replication is paramount to identifying targets for intervention. Here, we provide a brief summary of human gammaherpesvirus pathogenesis with a focus on advances made in the last 5 years.
Diseases Associated with Human Gammaherpesviruses
EBV-Associated Diseases
EBV was discovered in 1964 [1], and is implicated in many distinct diseases in both immunocompetent and immunocompromised individuals. Disease can arise from both epithelial and B-cell lineages. There are many manifestations of EBV-associated diseases [2]. EBV infection can result in non-malignant diseases such as infectious mononucleosis, and chronic active infection. EBV can also cause many different malignancies in immunocompetent hosts, including Burkitt and classical Hodgkin lymphoma in B cells, and extranodal natural killer (NK) NK/T cell and virus-associated hemophagocytic T cell lymphomas. EBV is also associated with epithelial cell malignancies including nasopharyngeal carcinoma and gastric cancer.
In immunocompromised hosts, disease manifestations are defined by whether the patient has congenital or acquired immunodeficiency. Congenital immunodeficiency can result in several B-cell lymphomas: severe combined immunodeficiency-associated, Wiskott-Aldrich syndrome, and X-linked lymphoproliferative disorder-associated. Acquired immunodeficiency due to organ transplantation or acquired immunodeficiency syndrome (AIDS) can result in multiple EBV-associated disease manifestations such as AIDS-associated and methotrexate-associated B-cell lymphomas, post-transplant lymphoproliferative disorder (PTLD), lymphomatoid granulomatosis, and oral hairy leukoplakia.
KSHV-Associated Disease
KSHV was discovered in 1994 [3], and is linked to four distinct disease manifestations [4]. KSHV is the etiological agent of the endothelial cancer Kaposi’s sarcoma and can be associated with AIDS-dependent and -independent malignancies. Primary effusion lymphoma is a KSHV-associated malignancy of B cells, which can also be co-infected with EBV [5]. KSHV is also associated with lymphoproliferative multicentric Castleman’s disease. Most recently, coinfection with human immunodeficiency virus (HIV) was found to be associated with KSHV-inflammatory cytokine syndrome, where high levels of viremia trigger a cytokine storm [6].
Recent Discoveries on Tropism Determinants of EBV and KSHV
Viral entry is the first step of infection. Since herpesviruses establish a lifelong infection, understanding the process of entry and fusion can aid in developing vaccines and inhibitor targets. All herpesviruses encode and utilize viral glycoproteins B (gB) and gH/gL for cellular entry. Understanding the additional proteins required for EBV and KSHV tropism is critical to establishing a mechanism of how entry and fusion occur.
Entry of EBV into epithelial cells requires gB, gH/gL, and an additional protein, BMRF, which binds β1 integrin [7, 8]. Within the past five years, receptor ephrin tyrosine kinase A2 (EphA2) was recently identified by two research groups as an essential epithelial cell receptor that binds gH/gL to drive fusion [9*, 10*]. Elucidating how EphA2 binds and activates gH/gL will help characterize the entry mechanism into epithelial cells. Attachment to B cells is mediated by EBV gp350, which binds to B cell-specific complement receptor 2 (CR2/CD21) [11]. Following attachment, EBV gp42 binds human leukocyte antigen (HLA)-II [12], triggering the fusion cascade. Fusion requires activated gp42, gB, and gH/gL. Recently, the structure of the gH/gL-gp42 complex was reported, in which an antibody against gH blocked EBV entry into epithelial cells, but not B cells, revealing the structural interfaces involved in the distinct entry mechanisms between the two cell types [13**]. Solving the gH/gL-gp42 crystal structure was crucial to understanding how gp42 acts as a determinant of EBV tropism, with high or low levels promoting entry into B cells or epithelial cells, respectively [14]. Intriguingly, the incorporation of gp42 is dependent on the cell type in which replication occurs, with B cells producing low abundance, and epithelial cells producing high abundance, gp42 virions, which is thought to help drive the tropism of EBV [14]. Advances in understanding the EBV fusion mechanism has led to the development of structurally designed gp350 and gH/gL-gp42 nanoparticles that are very promising vaccine candidates [15, 16**].
Relative to EBV, much less is known about KSHV entry and fusion. Several receptors have been previously identified for KSHV [17]. Additionally, during the past five years, it has been demonstrated that KSHV gH has an EphA2-binding site, and soluble EphA2 blocks entry, but mutating the binding site on gH has no effect on entry [18]. This could be explained by KSHV utilizing EphA4 [19, 20]. Compared to EBV, specific viral determinants of tropism have taken longer to identify. K8.1A is known to bind heparan sulfate [21] and was previously deemed dispensable for entry [22]. Recently, due to the development of a cell-free B cell infection system [23], K8.1A has been identified as indispensable for entry into B cells [24*]. These results suggest the role of K8.1A in B-cell entry should be further elucidated.
While significant progress has been made in understanding gammaherpesvirus tropism, especially in EBV, more work is needed to help model the mechanisms of entry and identify new viral and host determinants of entry. Whether K8.1A or another KSHV protein drives tropism in an EBV gp42-like mechanism is an intriguing question. Altogether, both EBV and KSHV have complex lifecycles, with specific tropism determinants that help drive viral pathogenesis.
Establishment and Maintenance of Latency
Establishing latency is a critical aspect of herpesvirus persistence in the population and in the host. Evading host detection by modulating cell pathways and expressing a limited repertoire of viral transcripts has allowed gammaherpesviruses to succeed as pathogens. Understanding how EBV and KSHV establish and maintain latency is an important biological question. EBV and KSHV produce very few transcripts during latency compared to during lytic replication, and expression profiles can be cell-type dependent [25–28]. EBV and KSHV encode two types of transcripts: those that code for proteins that antagonize cellular responses and modulate pathways to promote viral latency, and non-coding RNA that downregulate specific host cell proteins through RNAi.
EBV Latency-Associated Transcripts
Expression of latent EBV transcripts is very cell-type dependent, with distinct expression profiles and resulting types of latency I,II and III [29]. Cells displaying type I latency express EBV nuclear antigen (EBNA)-1, latent membrane protein (LMP)-2a/b, and EBV-encoded small RNAs (EBERS). During type II latency, EBNA-1/2, LMP-1, LMP-2a/b, and EBERs are expressed. Type III latency is characterized by the expression of the majority of the latent genes; EBNA-1, −2, −3a, −3b, −3c, and -LP (leader protein), LMP-1, −2a, −2b, and EBERs [30].
The EBNAs are largely thought to be important in viral genome segregation during cell division and regulation of viral and host transcription [30]. LMP-1 is critical in modulating host pathways, specifically the nuclear factor-kappa B (NF-κB) cascade [31]. LMP-2a mimics host B-cell receptor (BCR) signaling to prevent antigen recognition, among other modulatory roles [32]. EBV miRNAs are hypothesized to modulate host pathways to establish and maintain latency [33, 34]. The successful establishment and maintenance of EBV latency are modulated by several transcripts that have distinct and important roles in modifying and evading the host.
KSHV Latency-Associated Transcripts
KSHV transcripts expressed during latency are also dependent on cell type; generally, latency-associated nuclear antigen (LANA), vCyclin, vFLIP, viral miRNAs, kaposin, and vIRF3 (also known as LANA2) are expressed. There is evidence that viral interleukin (vIL6), K1, and K15 are expressed at low levels in some cells [35, 36]. LANA is critical in establishing latency through its ability to regulate viral and host transcription and to interact with many host proteins. One of LANA’s most important roles is maintaining the viral episomal genome and ensuring proper genome segregation during cellular division [37]. vCyclin is important in regulating cell cycle progression [38]. vFLIP has been identified as an integral protein in activating NF-κB to prevent apoptosis [39]. Viral miRNAs target multiple host proteins to modulate the host environment and maintain latency [40]. Differential translation initiation results in multiple transcripts produced by the kaposin locus that are involved in transformation and increasing cytokine expression [41, 42]. vIRF3 plays an important role in disrupting interferon response to latent infection [43, 44]. Finally, K15 has recently been shown to co-localize with phospholipase C γ1 (PLCγI) in latent cells which may drive angiogenesis and invasiveness [45]. In sum, similarly to EBV, KSHV encodes a small but specific suite of transcripts to promote and maintain viral latency.
Functional Homology of Human Gammaherpesvirus Latency Transcripts
While EBV and KSHV express distinct non-homologous transcripts during latency, there is functional homology between the subsets of genes expressed by both viruses. The EBNAs and LANA regulate viral and host transcription, aiding in genome segregation and in binding host cell proteins to promote infection by EBV and KSHV, respectively. Among other roles, EBV LMP-1 and KSHV vFLIP activate NF-κB to prevent host cell apoptosis. Finally, it has recently become very evident that EBV and KSHV encode and utilize non-coding RNAs to alter the host to their advantage. Understanding whether EBV and KSHV modulate similar pathways with these RNAs is an important question.
Advances in Understanding How EBV and KSHV Modulate the Host Environment:
In order to persist in the host, both EBV and KSHV must alter the host environment to be permissive to latency. Gammaherpesviruses modulate many pathways to ensure persistence of latently-infected cells. Both KSHV and EBV antagonize host p53 to prevent apoptosis [25, 46], and additional KSHV-encoded binding partners of p53 were recently identified [47]. KSHV epigenetically downregulates host cell PDZ and LIM Domain 2 (PDLIM2) to prevent the degradation of NF-κB [48]; whether EBV regulates PDLIM2 in a similar manner is unclear at this time.
Kinases regulate many host pathways, and unsurprisingly, KSHV and EBV both utilize and modulate kinase pathways for survival and pathogenesis. EBV and KSHV activate the phosphatidylinositol-3-kinase/AKT/mammalian target of rapamycin (PI3K/AKT/mTOR) pathways for cell survival via LMP-1 and LMP-2a [49, 50] or vIL6, vGPCR, and K1, respectively [51]. Both KSHV and EBV activate the Janus kinase/signal transducers and activators of transcription (JAK/STAT) pathway to ensure cell survival by vIL6 [52] and LMP1 [53], respectively.
In addition to changes mediated by viral proteins, EBV and KSHV encode miRNAs that are hypothesized to regulate the host to be amenable to infection [54–56]. EBV miRNA stabilizes latency by downregulating lytic transcription [57–59]. EBV downregulates host miRNAs and proteins to modulate the host cell environment to be conducive to latency [58–61]. The entire KSHV miRNA cluster modulates the host metabolic response by downregulating hypoxia-inducible factor (HIF), prolyl hydroxylase EGLN2, and heat shock protein A 9 (HSPA9) [62]. KSHV miRNA also maintains nitric oxide (NO) production, which is essential for the survival of KSHV-infected and transformed cells [63]. Recently, KSHV extracellular vesicles were found to modulate neighboring cells without triggering innate immune responses [64*]; it is possible this is accomplished similarly to EBV transmitting miRNAs to neighboring cells [65].
To summarize, it is increasingly evident that human gammaherpesviruses significantly alter the host cell in order to establish long-term latency.
Reactivation From Latency
Both KSHV and EBV preferentially establish latency during primary infection, which makes studying lytic replication challenging. For this reason, the lytic stage of the lifecycle is often studied in the context of viral reactivation from latency. Studying reactivation from latency is crucial. In order to complete the herpesviral life cycle, lytic infection needs to proceed to produce virions to infect new cells and new hosts. Additionally, in EBV and KSHV-associated cancers, there are detectable lytic transcripts, suggesting their importance in oncogenesis. For EBV and KSHV, there is a balancing act between maintaining latency and reactivating from latency. EBNA inhibits viral BRLF1 and BZLF1 expression while LANA inhibits replication and transcription activator (RTA) expression and vice versa for both EBV and KSHV, respectively. The complex interactions required to establish and maintain latency have resulted in identifying many distinct cellular factors that regulate reactivation from latency.
Cell Factor Dependent Reactivation
Cellular differentiation has long been thought to be a trigger of EBV and KSHV reactivation from latency. Host cell differentiation factor B-lymphocyte-induced maturation protein 1 (BLIMP1) activates EBV lytic transcription, and knockout of BLIMP1 results in less lytic gene production [66–68]. It has not been determined if BLIMP1 activates KSHV reactivation. Epithelial cell exosomes can trigger EBV reactivation of B cells [69*]. It is possible that these exosomes, coupled with differential glycoprotein repertoires expressed in virions from distinct cell types, drive EBV tropism.
Episomal Modification
Due to the limited repertoire of proteins expressed by EBV and KSHV during latency, it is unsurprising that many restrictors of reactivation from latency are associated directly with viral episomes. Modification of KSHV [70*, 71*] and EBV [72] episomes can regulate reactivation from latency. By directly binding RTA, it was recently shown that fused in sarcoma (FUS) restricts KSHV reactivation from latency [73]. PARP1 restricts both KSHV [74] and EBV reactivation [75] by binding RTA and BZLF, respectively. Disruption of Kruppel-associated box domain-associated protein-1 (KAP1) binding to viral episomes results in reactivation of KSHV [76] and EBV [77].
Innate Immune Restriction
The nature of latent infection limits the ability of the host adaptive immune system to identify infected cells, making the innate immune response critical in restricting reactivation and lytic replication. DNA sensors cyclic guanosine monophosphate-adenosine monophosphate synthase (cGAS) and stimulator of interferon genes (STING) have recently been shown to regulate KSHV reactivation from latency [78–80], but whether this holds true in the EBV lifecycle has not yet been determined. EBV reactivation has been shown to activate RNA sensor retinoic acid-inducible gene I (RIG-I ) [81], and KSHV reactivation was found to also trigger RIG-I to induce interferon [82*–84]. Over the last five years, interferon-induced protein with tetratricopeptide repeats (IFIT) 1 and 3 [85] and interferon stimulated gene 15 (ISG15) [86] have been found to restrict KSHV reactivation, while Nod-like receptor X1 (NLRX1) negatively regulates reactivation from latency [87]. Further studies into how the innate immune system restricts KSHV and EBV are integral to understand reactivation from latency.
Kinase Regulated Reactivation
Kinases play an important role throughout the KSHV lifecycle, and reactivation from latency is no exception. For example, knocking down all known human kinases identified tousled-like kinase 2 (TLK2) as a restrictor against reactivation, and this was subsequently validated in EBV [88]. Mitogen-activated protein kinase (MAPK) restricts KSHV [32, 89] and EBV [90] reactivation. Kinases are popular drug candidates in human medicine and perhaps could become attractive targets in treatment of gammaherpesvirus infection [91].
Altogether, human gammaherpesviruses reactivation is regulated by many host and viral factors. Reactivation from latency is a critical step to drive lytic replication and produce new infectious progeny.
Lytic Replication
EBV and KSHV lytic replication following reactivation is essential to produce new infectious virions. Lytic replication is largely conserved throughout all herpesviruses. Briefly, immediate early (IE) gene expression is required for full lytic reactivation and is regulated through a feed forward mechanism in which RTA in KSHV and BRLF1/BZLF1 in EBV act as transcription activators for the other IE genes. As transcriptional activators, IE proteins activate delayed early (DE) gene transcription in a DNA replication-independent mechanism. Early genes encode proteins involved in the DNA replication machinery. After the initiation of viral DNA replication, the late genes are transcribed, which mostly encode structural proteins. Viral DNA-containing capsids are assembled within, and then trafficked out of, the nucleus. In the cytoplasm, capsids obtain most tegument proteins and the viral lipid bilayer. From there, the new infectious progeny is produced and released from the cell, ready to infect a new cell or host.
KSHV Lytic Replication
KSHV replication is activated by RTA. KSHV is unique in that it encodes multiple proteins that were likely repurposed from the host cell and act as homologs, including vIL6, G protein-coupled receptor (vGPCR), protein kinase (vPK), and the interferon regulatory factors (vIRFs). KSHV utilizes these host-derived homologs to productively influence cell signaling pathways to its benefit. Additionally, KSHV also encodes genes that are not homologous to cellular genes, e.g., K1 and K15.
vGPCR, vIL6, and K1 can prevent cell death by activating pathways such as PI3K/AKT/mTOR [51]. K1 also modulates AMP-activated protein kinase (AMPK) to promote cell survival [92]. vIRF1, −2, and −4 antagonize interferon response to aid in innate immune avoidance [44, 93, 94]. vIL6, like its host homolog IL6, activates the JAK/STAT signaling pathway [52]. KSHV ORFs have many functions beyond those listed here, and there are likely still unknown functions to identify. Non-coding polyadenylated nuclear (PAN) RNA is one of the most abundant transcripts and performs a wide variety of functions [95]. circRNA has been identified during KSHV lytic replication, and its function remains to be elucidated [96**, 97**].
EBV Lytic Replication
Compared to KSHV, EBV has a limited number of unique proteins that aid in lytic replication. Distinct from KSHV RTA, EBV uses two transcriptional activators to drive lytic gene transcription: BZLF1 and BRLF1, which depend on the cellular context. EBV viral kinase BGLF2 activates c-Jun N-terminal kinase (JNK) and MAPK, which enhances BZLF1 expression to drive lytic replication [98]. Cellular helicase DHX9, a member of the DExD/H-box family, restricts EBV lytic replication by binding to viral protein SM [99]. circRNA has been identified during EBV lytic replication, but its role in replication remains unclear [97**, 100**].
Collectively, KSHV and EBV modulate multiple host pathways, particularly kinases, to facilitate lytic replication to produce new virions.
Conclusions
EBV and KSHV are the etiological agents of several cancers worldwide. There are no FDA-approved vaccines for either virus. They effectively persist in the human population by utilizing distinct protein expression profiles in different cell types at different stages of their life cycles. A model depicting the gammaherpesvirus lifecycle is shown in Fig. 1. The recent discovery of EBV glycoprotein-receptor crystal structures, has advanced the field. Identifying how non-coding RNAs modulate the host’s susceptibility to infection is important throughout the life cycle, and this work will continue to reveal new details of KSHV and EBV’s replication. Both viral and host kinases have significant roles in multiple steps of the EBV and KSHV life cycle and may have therapeutic potential. Much has been accomplished in understanding host restriction of reactivation from latency, specifically how episomal modification of viral genomes triggers lytic replication. Unraveling how KSHV and EBV establish and reactivate from latency and proceed to lytic infection is a broad question that will continue to advance understanding of viral pathogenesis.
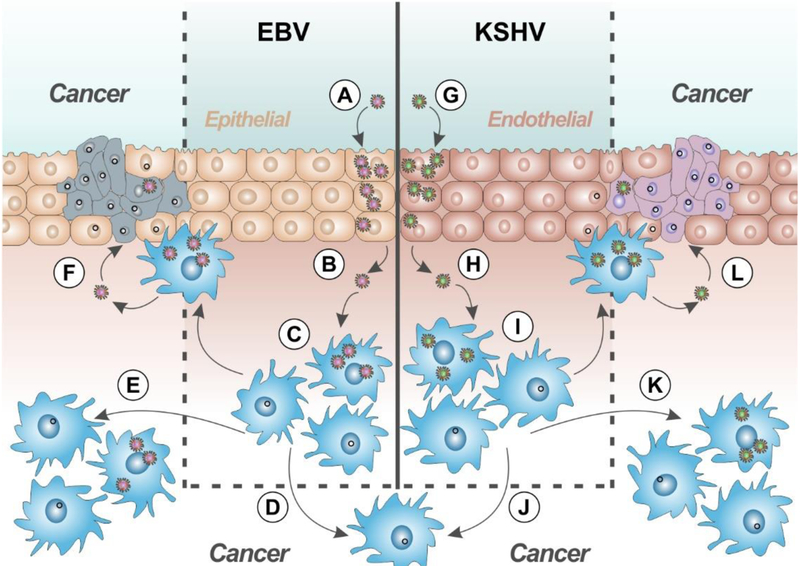
Figure 1.
Proposed model for human gammaherpesvirus pathogenesis. Primary infection of epithelial cells by EBV (A) and endothelial cells by KSHV (G). Released virions then infect lymphocytes (B-C, H-I). Lymphocytes infected with KSHV +/− EBV may result in primary effusion lymphoma (D,J). EBV-infected cells are associated with lymphatic (E) and epithelial cancers (F). KSHV-infected cells are associated with lymphatic (K) and endothelial (L) cancers.
Acknowledgments
We apologize for those whose work we could not cite due to space restraints and also due to the restriction on mainly citing literature in the past five years. We thank Damania lab members for helpful edits. We are extremely grateful to Rebecca Weed for creation of the figure.
Funding Information
This work was supported by public health service grants CA096500 (BD), CA019014 (BD), DE028211 (BD), CA163217 (BD), and T32CA009156 (DJW). BD is a Burroughs Wellcome Fund Investigator in Infectious Disease and a Leukemia and Lymphoma Society Scholar.
References
- 1.Epstein MA, Achong BG, Barr YM. Virus particles in cultured lymphoblasts from Burkitt’s lymphoma. The Lancet. 1964;283(7335):702–3. [DOI] [PubMed] [Google Scholar]
- 2.Kutok J, Wang F. Spectrum of Epstein-Barr virus-associated diseases. Annu Rev Pathol Mech Dis. 2006;1:375–404. [DOI] [PubMed] [Google Scholar]
- 3.Chang Y, Cesarman E, Pessin MS, Lee F, Culpepper J, Knowles DM et al. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi’s sarcoma. Science. 1994;266(5192):1865–9. [DOI] [PubMed] [Google Scholar]
- 4.Dittmer DP, Damania B. Kaposi sarcoma associated herpesvirus pathogenesis (KSHV)—an update. Current opinion in virology. 2013;3(3):238–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klepfish A, Sarid R, Shtalrid M, Shvidel L, Berrebi A, Schattner A. Primary effusion lymphoma (PEL) in HIV-negative patients-a distinct clinical entity. Leukemia & lymphoma. 2001;41(3–4):439–43. [DOI] [PubMed] [Google Scholar]
- 6.Polizzotto MN, Uldrick TS, Wyvill KM, Aleman K, Marshall V, Wang V et al. Clinical features and outcomes of patients with symptomatic Kaposi sarcoma herpesvirus (KSHV)-associated inflammation: prospective characterization of KSHV inflammatory cytokine syndrome (KICS). Clinical Infectious Diseases. 2015;62(6):730–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xiao J, Palefsky JM, Herrera R, Tugizov SM. Characterization of the Epstein-Barr virus glycoprotein BMRF-2. Virology. 2007;359(2):382–96. [DOI] [PubMed] [Google Scholar]
- 8.Xiao J, Palefsky JM, Herrera R, Berline J, Tugizov SM. The Epstein-Barr virus BMRF-2 protein facilitates virus attachment to oral epithelial cells. Virology. 2008;370(2):430–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.*.Zhang H, Li Y, Wang H-B, Zhang A, Chen M-L, Fang Z-X et al. Ephrin receptor A2 is an epithelial cell receptor for Epstein-Barr virus entry. Nature microbiology. 2018;3(2):164. [DOI] [PubMed] [Google Scholar]; These two papers established Ephrin A2 as an entry receptor for EBV
- 10.*.Chen J, Sathiyamoorthy K, Zhang X, Schaller S, White BEP, Jardetzky TS et al. Ephrin receptor A2 is a functional entry receptor for Epstein-Barr virus. Nature microbiology. 2018;3(2):172. [DOI] [PMC free article] [PubMed] [Google Scholar]; These two papers established Ephrin A2 as an entry receptor for EBV
- 11.Lowell CA, Klickstein LB, Carter RH, Mitchell JA, Fearon DT, Ahearn J. Mapping of the Epstein-Barr virus and C3dg binding sites to a common domain on complement receptor type 2. Journal of Experimental Medicine. 1989;170(6):1931–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mullen MM, Haan KM, Longnecker R, Jardetzky TS. Structure of the Epstein-Barr virus gp42 protein bound to the MHC class II receptor HLA-DR1. Molecular cell. 2002;9(2):375–85. [DOI] [PubMed] [Google Scholar]
- 13.**.Sathiyamoorthy K, Hu YX, Mohl BS, Chen J, Longnecker R, Jardetzky TS. Structural basis for Epstein- Barr virus host cell tropism mediated by gp42 and gHgL entry glycoproteins. Nature communications. 2016;7:13557. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper crystalized the gp42/gH/gL complex, yielding valuable structural insights into EBV fusion
- 14.Borza CM, Hutt-Fletcher LM. Alternate replication in B cells and epithelial cells switches tropism of Epstein-Barr virus. Nature medicine. 2002;8(6):594. [DOI] [PubMed] [Google Scholar]
- 15.Kanekiyo M, Bu W, Joyce MG, Meng G, Whittle JR, Baxa U et al. Rational design of an Epstein-Barr virus vaccine targeting the receptor-binding site. Cell. 2015;162(5):1090–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.**.Bu W, Joyce MG, Nguyen H, Banh DV, Aguilar F, Tariq Z et al. Immunization with Components of the Viral Fusion Apparatus Elicits Antibodies That Neutralize Epstein-Barr Virus in B Cells and Epithelial Cells. Immunity. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]; This work develops and characterizes a promising EBV vaccine candidate
- 17.Kumar B, Chandran B. KSHV entry and trafficking in target cells—hijacking of cell signal pathways, actin and membrane dynamics. Viruses. 2016;8(11):305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Großkopf AK, Ensser A, Neipel F, Jungnickl D, Schlagowski S, Desrosiers RC et al. A conserved Eph family receptor-binding motif on the gH/gL complex of Kaposi’s sarcoma-associated herpesvirus and rhesus monkey rhadinovirus. PLoS pathogens. 2018;14(2):e1006912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.TerBush AA, Hafkamp F, Lee HJ, Coscoy L. A Kaposi’s Sarcoma-Associated Herpesvirus Infection Mechanism is Independent of Integrins a3p i, aVP3, and aVP5. Journal of virology. 2018:JVI. 00803–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen J, Zhang X, Schaller S, Jardetzky TS, Longnecker R. Ephrin Receptor A4 is a New Kaposi’s Sarcoma-Associated Herpesvirus Virus Entry Receptor. mBio. 2019;10(1):e02892–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Birkmann A, Mahr K, Ensser A, Yaguboglu S, Titgemeyer F, Fleckenstein B et al. Cell surface heparan sulfate is a receptor for human herpesvirus 8 and interacts with envelope glycoprotein K8. 1. Journal of virology. 2001;75(23):11583–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luna RE, Zhou F, Baghian A, Chouljenko V, Forghani B, Gao S-J et al. Kaposi’s sarcoma-associated herpesvirus glycoprotein K8. 1 is dispensable for virus entry. Journal of virology. 2004;78(12):6389–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dollery SJ, Santiago-Crespo RJ, Kardava L, Moir S, Berger EA. Efficient infection of a human B cell line with cell-free Kaposi’s sarcoma-associated herpesvirus. Journal of virology. 2014;88(3):1748–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.*.Dollery SJ, Santiago-Crespo RJ, Chatterjee D, Berger EA. Glycoprotein K8. 1A of Kaposi’s sarcoma-associated herpesvirus is a critical B cell tropism determinant, independent of its heparan sulfate binding activity. Journal of virology. 2018:JVI. 01876–18. [DOI] [PMC free article] [PubMed] [Google Scholar]; This is the first report of a tropism determinant of KSHV entry
- 25.Rivas C, Thlick A-E, Parravicini C, Moore PS, Chang Y. Kaposi’s sarcoma-associated herpesvirus LANA2 is a B-cell-specific latent viral protein that inhibits p53. Journal of virology. 2001;75(1):429–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dittmer DP. Transcription profile of Kaposi’s sarcoma-associated herpesvirus in primary Kaposi’s sarcoma lesions as determined by real-time PCR arrays. Cancer research. 2003;63(9):2010–5. [PubMed] [Google Scholar]
- 27.Kerr B, Lear A, Rowe M, Croom-Carter D, Young L, Rookes S et al. Three transcriptionally distinct forms of Epstein-Barr virus latency in somatic cell hybrids: cell phenotype dependence of virus promoter usage. Virology. 1992;187(1):189–201. [DOI] [PubMed] [Google Scholar]
- 28.Miyashita EM, Yang B, Babcock GJ, Thorley-Lawson DA. Identification of the site of Epstein-Barr virus persistence in vivo as a resting B cell. Journal of virology. 1997;71(7):4882–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rowe M, Lear A, Croom-Carter D, Davies A, Rickinson A. Three pathways of Epstein-Barr virus gene activation from EBNA1-positive latency in B lymphocytes. Journal of virology. 1992;66(1):122–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kang M-S, Kieff E. Epstein-Barr virus latent genes. Experimental & molecular medicine. 2015;47(1):e131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huen D, Henderson S, Croom-Carter D, Rowe M. The Epstein-Barr virus latent membrane protein-1 (LMP1) mediates activation of NF-kappa B and cell surface phenotype via two effector regions in its carboxy-terminal cytoplasmic domain. Oncogene. 1995;10(3):549–60. [PubMed] [Google Scholar]
- 32.Caldwell RG, Wilson JB, Anderson SJ, Longnecker R. Epstein-Barr virus LMP2A drives B cell development and survival in the absence of normal B cell receptor signals. Immunity. 1998;9(3):405–11. [DOI] [PubMed] [Google Scholar]
- 33.Pfeffer S, Zavolan M, Grasser FA, Chien M, Russo JJ, Ju J et al. Identification of virus-encoded microRNAs. Science. 2004;304(5671):734–6. [DOI] [PubMed] [Google Scholar]
- 34.Skalsky RL, Cullen BR. EBV noncoding RNAs Epstein Barr Virus Volume 2 Springer; 2015. p. 181–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chandriani S, Ganem D. Array-based transcript profiling and limiting-dilution reverse transcription PCR analysis identify additional latent genes in Kaposi’s sarcoma-associated herpesvirus. Journal of virology. 2010;84(11):5565–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sharp TV, Wang H-W, Koumi A, Hollyman D, Endo Y, Ye H et al. K15 protein of Kaposi’s sarcoma-associated herpesvirus is latently expressed and binds to HAX-1, a protein with antiapoptotic function. Journal of virology. 2002;76(2):802–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Uppal T, Banerjee S, Sun Z, Verma S, Robertson E. KSHV LANA—the master regulator of KSHV latency. Viruses. 2014;6(12):4961–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Verschuren EW, Jones N, Evan GI. The cell cycle and how it is steered by Kaposi’s sarcoma-associated herpesvirus cyclin. Journal of General Virology. 2004;85(6):1347–61. [DOI] [PubMed] [Google Scholar]
- 39.Matta H, Chaudhary PM. Activation of alternative NF-k B pathway by human herpes virus 8-encoded Fas-associated death domain-like IL-ip-converting enzyme inhibitory protein (vFLIP). Proceedings of the National Academy of Sciences. 2004;101(25):9399–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Samols MA, Skalsky RL, Maldonado AM, Riva A, Lopez MC, Baker HV et al. Identification of cellular genes targeted by KSHV-encoded microRNAs. PLoS pathogens. 2007;3(5):e65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Muralidhar S, Pumfery AM, Hassani M, Sadaie MR, Azumi N, Kishishita M et al. Identification of kaposin (open reading frame K12) as a human herpesvirus 8 (Kaposi’s sarcoma-associated herpesvirus) transforming gene. Journal of virology. 1998;72(6):4980–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McCormick C, Ganem D. The kaposin B protein of KSHV activates the p38/MK2 pathway and stabilizes cytokine mRNAs. Science. 2005;307(5710):739–41. [DOI] [PubMed] [Google Scholar]
- 43.Lubyova B, Pitha PM. Characterization of a novel human herpesvirus 8-encoded protein, vIRF-3, that shows homology to viral and cellular interferon regulatory factors. Journal of virology. 2000;74(17):8194–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gao S-J, Boshoff C, Jayachandra S, Weiss RA, Chang Y, Moore PS. KSHV ORF K9 (vIRF) is an oncogene which inhibits the interferon signaling pathway. Oncogene. 1997;15(16):1979. [DOI] [PubMed] [Google Scholar]
- 45.Gramolelli S, Weidner-Glunde M, Abere B, Viejo-Borbolla A, Bala K, Ruckert J et al. Inhibiting the recruitment of PLCyl to Kaposi’s sarcoma herpesvirus K15 protein reduces the invasiveness and angiogenesis of infected endothelial cells. PLoS pathogens. 2015;11(8):e1005105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Okan I, Wang Y, Chen F, Hu L-F, Imreh S, Klein G et al. The EBV-encoded LMP1 protein inhibits p53-triggered apoptosis but not growth arrest. Oncogene. 1995;11(6):1027–31. [PubMed] [Google Scholar]
- 47.Chudasama P, Konrad A, Jochmann R, Lausen B, Holz P, Naschberger E et al. Structural proteins of Kaposi’s sarcoma-associated herpesvirus antagonize p53-mediated apoptosis. Oncogene. 2015;34(5):639. [DOI] [PubMed] [Google Scholar]
- 48.Sun F, Xiao Y, Qu Z. Oncovirus Kaposi sarcoma herpesvirus (KSHV) represses tumor suppressor PDLIM2 to persistently activate nuclear factor k B (NF-k B) and STAT3 transcription factors for tumorigenesis and tumor maintenance. Journal of Biological Chemistry. 2015;290(12):7362–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Portis T, Longnecker R. Epstein-Barr virus (EBV) LMP2A mediates B-lymphocyte survival through constitutive activation of the Ras/PI3K/Akt pathway. Oncogene. 2004;23(53):8619. [DOI] [PubMed] [Google Scholar]
- 50.Shair KH, Bendt KM, Edwards RH, Bedford EC, Nielsen JN, Raab-Traub N. EBV latent membrane protein 1 activates Akt, NFkB, and Stat3 in B cell lymphomas. PLoS pathogens. 2007;3(11):e166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bhatt AP, Damania B. AKTivation of PI3K/AKT/mTOR signaling pathway by KSHV. Frontiers in immunology. 2013;3:401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Molden J, Chang Y, You Y, Moore PS, Goldsmith MA. A Kaposi’s sarcoma-associated herpesvirus-encoded cytokine homolog (vIL-6) activates signaling through the shared gp130 receptor subunit. Journal of Biological Chemistry. 1997;272(31):19625–31. [DOI] [PubMed] [Google Scholar]
- 53.Gires O, Kohlhuber F, Kilger E, Baumann M, Kieser A, Kaiser C et al. Latent membrane protein 1 of Epstein-Barr virus interacts with JAK3 and activates STAT proteins. The EMBO journal. 1999;18(11):3064–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cai X, Lu S, Zhang Z, Gonzalez CM, Damania B, Cullen BR. Kaposi’s sarcoma-associated herpesvirus expresses an array of viral microRNAs in latently infected cells. Proceedings of the National Academy of Sciences. 2005;102(15):5570–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cai X, Schafer A, Lu S, Bilello JP, Desrosiers RC, Edwards R et al. Epstein-Barr virus microRNAs are evolutionarily conserved and differentially expressed. PLoS pathogens. 2006;2(3):e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Samols MA, Hu J, Skalsky RL, Renne R. Cloning and identification of a microRNA cluster within the latency-associated region of Kaposi’s sarcoma-associated herpesvirus. Journal of virology. 2005;79(14):9301–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jung Y-J, Choi H, Kim H, Lee SK. MicroRNA miR-BART20–5p stabilizes Epstein-Barr virus latency by directly targeting BZLF1 and BRLF1. Journal of virology. 2014;88(16):9027–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Haar J, Contrant M, Bernhardt K, Feederle R, Diederichs S, Pfeffer S et al. The expression of a viral microRNA is regulated by clustering to allow optimal B cell transformation. Nucleic acids research. 2015;44(3):1326–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Marquitz AR, Raab-Traub N, editors. The role of miRNAs and EBV BARTs in NPC Seminars in cancer biology; 2012: Elsevier. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mansouri S, Pan Q, Blencowe BJ, Claycomb JM, Frappier L. Epstein-Barr virus EBNA1 protein regulates viral latency through effects on let-7 microRNA and dicer. Journal of virology. 2014;88(19):11166–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Qiu J, Thorley-Lawson DA. EBV microRNA BART 18–5p targets MAP3K2 to facilitate persistence in vivo by inhibiting viral replication in B cells. Proceedings of the National Academy of Sciences. 2014;111(30):11157–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yogev O, Lagos D, Enver T, Boshoff C. Kaposi’s sarcoma herpesvirus microRNAs induce metabolic transformation of infected cells. PLoS pathogens. 2014;10(9):e1004400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li T, Zhu Y, Cheng F, Lu C, Jung JU, Gao S-J. Oncogenic Kaposi’s sarcoma-associated herpesvirus upregulates argininosuccinate synthase 1, a rate-limiting enzyme of the citrulline-nitric oxide cycle, to activate the STAT3 pathway and promote growth transformation. Journal of Virology. 2019;93(4):e01599–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.*.McNamara RP, Chugh PE, Bailey A, Costantini LM, Ma Z, Bigi R et al. Extracellular vesicles from Kaposi Sarcoma-associated herpesvirus lymphoma induce long-term endothelial cell reprogramming. PLoS pathogens. 2019;15(2):e1007536. [DOI] [PMC free article] [PubMed] [Google Scholar]; This is the first report of exosomes produced during KSHV infection and their role
- 65.Pegtel DM, Cosmopoulos K, Thorley-Lawson DA, van Eijndhoven MA, Hopmans ES, Lindenberg JL et al. Functional delivery of viral miRNAs via exosomes. Proceedings of the National Academy of Sciences. 2010;107(14):6328–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nawandar DM, Ohashi M, Djavadian R, Barlow E, Makielski K, Ali A et al. Differentiation-dependent LMP1 expression is required for efficient lytic EBV reactivation in epithelial cells. Journal of virology. 2017:JVI. 02438–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nawandar DM, Wang A, Makielski K, Lee D, Ma S, Barlow E et al. Differentiation-dependent KLF4 expression promotes lytic Epstein-Barr virus infection in epithelial cells. PLoS pathogens. 2015;11(10):e1005195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Reusch JA, Nawandar DM, Wright KL, Kenney SC, Mertz JE. Cellular differentiation regulator BLIMP1 induces Epstein-Barr virus lytic reactivation in epithelial and B cells by activating transcription from both the R and Z promoters. Journal of virology. 2015;89(3):1731–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.*.Lin Z, Swan K, Zhang X, Cao S, Brett Z, Drury S et al. Secreted oral epithelial cell membrane vesicles induce Epstein-Barr virus (EBV) reactivation in latently infected B-cells. Journal of virology. 2016:JVI. 02830–15. [DOI] [PMC free article] [PubMed] [Google Scholar]; This work demonstrates host cell vesicles drive reactivation and may have a role in driving viral tropism
- 70.*.Hopcraft SE, Pattenden SG, James LI, Frye S, Dittmer DP, Damania B. Chromatin remodeling controls Kaposi’s sarcoma-associated herpesvirus reactivation from latency. PLoS pathogens. 2018;14(9):e1007267. [DOI] [PMC free article] [PubMed] [Google Scholar]; This work demonstrated the role chromatin remodeling has in controling KSHV reactivation latency
- 71.*.Hu M, Armstrong N, Seto E, Li W, Zhu F, Wang PC et al. Sirtuin 6 Attenuates Kaposi’s Sarcoma-associated herpesvirus (KSHV) Reactivation via Suppressing the Ori-Lyt Activity and Expression of RTA. Journal of virology. 2019:JVI. 02200–18. [DOI] [PMC free article] [PubMed] [Google Scholar]; This work showed that host Sirtuin 6 is an important regulator of KSHV reactivation
- 72.Murata T, Tsurumi T. Epigenetic modification of the Epstein-Barr virus BZLF1 promoter regulates viral reactivation from latency. Frontiers in genetics. 2013;4:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dunker W, Song Y, Zhao Y, Karijolich J. FUS Negatively Regulates Kaposi’s Sarcoma-Associated Herpesvirus Gene Expression. Viruses. 2018;10(7):359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gwack Y, Nakamura H, Lee SH, Souvlis J, Yustein JT, Gygi S et al. Poly (ADP-ribose) polymerase 1 and Ste20-like kinase hKFC act as transcriptional repressors for gamma-2 herpesvirus lytic replication. Molecular and cellular biology. 2003;23(22):8282–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lupey-Green LN, Moquin SA, Martin KA, McDevitt SM, Hulse M, Caruso LB et al. PARP1 restricts Epstein Barr Virus lytic reactivation by binding the BZLF1 promoter. Virology. 2017;507:220–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chang P-C, Fitzgerald LD, Van Geelen A, Izumiya Y, Ellison TJ, Wang D-H et al. Kruppel-associated box domain-associated protein-1 as a latency regulator for Kaposi’s sarcoma-associated herpesvirus and its modulation by the viral protein kinase. Cancer research. 2009;69(14):5681–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bentz GL, Moss CR, Whitehurst CB, Moody CA, Pagano JS. LMPl-induced Sumoylation Influences the Maintenance of EBV Latency Through KAP1. Journal of virology. 2015:JVI 00711–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ma Z, Jacobs SR, West JA, Stopford C, Zhang Z, Davis Z et al. Modulation of the cGAS-STING DNA sensing pathway by gammaherpesviruses. Proceedings of the National Academy of Sciences. 2015;112(31):E4306–E15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang G, Chan B, Samarina N, Abere B, Weidner-Glunde M, Buch A et al. Cytoplasmic isoforms of Kaposi sarcoma herpesvirus LANA recruit and antagonize the innate immune DNA sensor cGAS. Proceedings of the National Academy of Sciences. 2016;113(8):E1034–E43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wu J-j, Li W, Shao Y, Avey D, Fu B, Gillen J et al. Inhibition of cGAS DNA sensing by a herpesvirus virion protein. Cell host & microbe. 2015;18(3):333–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chiu Y-H, MacMillan JB, Chen ZJ. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell. 2009;138(3):576–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.*.West JA, Wicks M, Gregory SM, Chugh P, Jacobs SR, Zhang Z et al. An important role for mitochondrial antiviral signaling protein in the Kaposi’s sarcoma-associated herpesvirus life cycle. Journal of virology. 2014;88(10):5778–87. [DOI] [PMC free article] [PubMed] [Google Scholar]; This is the first manuscript that showed that the RNA sensor, RIG-I, and its adaptor MAVS can sense infection with a KSHV, a DNA virus.
- 83.Zhang Y, Dittmer DP, Mieczkowski PA, Host KM, Fusco WG, Duncan JA et al. RIG-I Detects Kaposi’s Sarcoma-Associated Herpesvirus Transcripts in a RNA Polymerase III-Independent Manner. mBio. 2018;9(4):e00823–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhao Y, Ye X, Dunker W, Song Y, Karijolich J. RIG-I like receptor sensing of host RNAs facilitates the cell-intrinsic immune response to KSHV infection. Nature communications. 2018;9(1):4841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Li D, Swaminathan S. Human IFIT proteins inhibit lytic replication of KSHV: A new feed-forward loop in the innate immune system. PLoS pathogens. 2019;15(2):e1007609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jacobs SR, Stopford CM, West JA, Bennett CL, Giffin L, Damania B. KSHV vIRF1 interacts with a member of the Interferon Stimulated Gene 15 pathway. Journal of virology. 2015:JVI 01482–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ma Z, Hopcraft SE, Yang F, Petrucelli A, Guo H, Ting JP et al. NLRX1 negatively modulates type I IFN to facilitate KSHV reactivation from latency. PLoS pathogens. 2017;13(5):e1006350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dillon PJ, Gregory SM, Tamburro K, Sanders MK, Johnson GL, Raab-Traub N et al. Tousled-like kinases modulate reactivation of gammaherpesviruses from latency. Cell host & microbe. 2013;13(2):204–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Haas DA, Bala K, Busche G, Weidner-Glunde M, Santag S, Kati S et al. The inflammatory kinase MAP4K4 promotes reactivation of Kaposi’s sarcoma herpesvirus and enhances the invasiveness of infected endothelial cells. PLoS pathogens. 2013;9(11):e1003737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Matusali G, Arena G, De Leo A, Di Renzo L, Mattia E. Inhibition of p38 MAP kinase pathway induces apoptosis and prevents Epstein Barr virus reactivation in Raji cells exposed to lytic cycle inducing compounds. Molecular cancer. 2009;8(1):18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wong JP, Stuhlmiller TJ, Giffin LC, Lin C, Bigi R, Zhao J et al. Kinome profiling of non-Hodgkin lymphoma identifies Tyro3 as a therapeutic target in primary effusion lymphoma. Proceedings of the National Academy of Sciences. 2019:201903991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Anders PM, Zhang Z, Bhende PM, Giffin L, Damania B. The KSHV K1 protein modulates AMPK function to enhance cell survival. PLoS pathogens. 2016;12(11):e1005985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Burysek L, Yeow W, Pitha P. Unique properties of a second human herpesvirus 8-encoded interferon regulatory factor (vIRF-2). Journal of human virology. 1999;2(1):19–32. [PubMed] [Google Scholar]
- 94.Hwang S-W, Kim D, Jung JU, Lee H-R. KSHV-encoded viral interferon regulatory factor 4 (vIRF4) interacts with IRF7 and inhibits interferon alpha production. Biochemical and biophysical research communications. 2017;486(3):700–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rossetto C, Pari G. PAN’s Labyrinth: Molecular biology of Kaposi’s sarcoma-associated herpesvirus (KSHV) PAN RNA, a multifunctional long noncoding RNA. Viruses. 2014;6(11):4212–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.**.Tagawa T, Gao S, Koparde VN, Gonzalez M, Spouge JL, Serquina AP et al. Discovery of Kaposi’s sarcoma herpesvirus-encoded circular RNAs and a human antiviral circular RNA. Proceedings of the National Academy of Sciences. 2018;115(50):12805–10. [DOI] [PMC free article] [PubMed] [Google Scholar]; This work identified and characterized circRNAs, which may prove to be very important in KSHVs lifecycle
- 97.**.Toptan T, Abere B, Nalesnik MA, Swerdlow SH, Ranganathan S, Lee N et al. Circular DNA tumor viruses make circular RNAs. Proceedings of the National Academy of Sciences. 2018;115(37):E8737–E45. [DOI] [PMC free article] [PubMed] [Google Scholar]; This manuscript described KSHV and EBV circRNAs, which is a new, novel finding
- 98.Liu X, Cohen JI. Epstein-Barr virus (EBV) tegument protein BGLF2 promotes EBV reactivation through activation of the p38 mitogen-activated protein kinase. Journal of virology. 2016;90(2):1129–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Fu W, Verma D, Burton A, Swaminathan S. Cellular RNA helicase DHX9 interacts with the essential Epstein-Barr virus (EBV) protein SM and restricts EBV lytic replication. Journal of virology. 2019;93(4):e01244–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.**.Ungerleider N, Concha M, Lin Z, Roberts C, Wang X, Cao S et al. The Epstein Barr virus circRNAome. PLoS pathogens. 2018;14(8):e1007206. [DOI] [PMC free article] [PubMed] [Google Scholar]; This manuscript describes EBV circRNAs as a new and novel discovery.