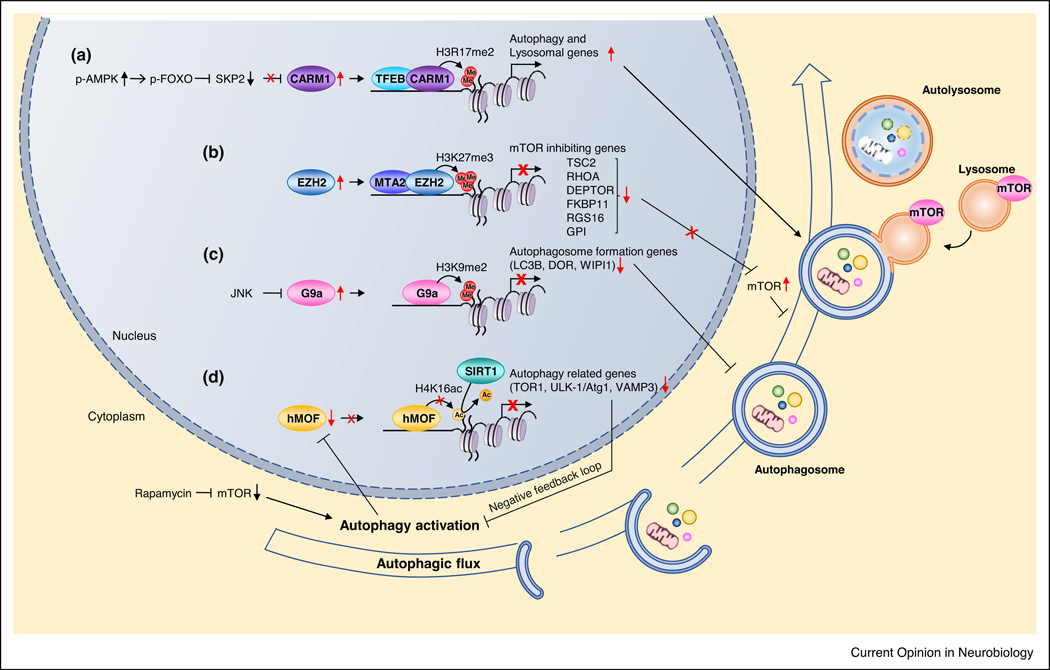
Figure 1.
Epigenetic regulators of autophagy genes. (a) CARM1 confers a dimethylation mark on H3 at arginine 17, which activates transcription of autophagy genes (ULK-1/Atg1, Atg11, Atg12) via the AMPK-SKP2-CARM1 signaling axis. (b) EZH-2 promotes trimethylation of H3 at lysine 27, and thereby represses genes that inhibit mTOR signaling (DEPTOR, TSC2) which in turn, activates mTOR signaling and suppresses autophagy. (c) G9a promotes demethylation of H3 at lysine 9, and thereby represses genes related to autophagosome formation. (d) Control of H4K16 acetylation status by hMOF and SIRT1. In response to cellular stress, mTOR (and Raptor) translocates to the surface of lysosomes, where mTOR is strategically positioned to be phosphorylated by AKT and to phosphorylate and inhibit its downstream target Transcription factor EB (TFEB), which drives transcription of genes crucial to autophagy and lysosomal degradation. Upon conditions of high nutrients, inhibition of mTOR signaling leads to activation of autophagy; under these conditions, hMOF and H4K16 acetylation are downregulated which in turn, represses transcription of autophagy related genes (TOR1, ULK-1/Atg1, VAMP3). Thus, a negative feedback regulatory loop occurs which might prevent overly high stimulation of autophagic flux.