Abstract
Salmonella is a poultry-borne pathogen that causes illness throughout the world. Consequently, it is critical to control Salmonella during the process of converting broilers to poultry meat. Sanitization of a poultry processing facility, including processing equipment, is a crucial control measure that is utilized by poultry integrators. However, prevalence of Salmonella on equipment after sanitization and its potential risk to food safety has not been evaluated thoroughly. Therefore, the objective of this study was to evaluate the persistence of Salmonella on poultry processing equipment before and following cleaning and sanitization procedure. A total of 15 locations within 6 commercial processing plants were sampled at 3 time points: (A) after processing; (B) after cleaning; and (C) after sanitization, on 3 separate visits for a total of 135 samples per plant. Salmonella-positive isolates were recovered from samples using the United States Department of Agriculture MLG 4.09 conventional method. Presumptive Salmonella colonies were subjected to biochemical tests for confirmation. Salmonella isolates recovered after sanitization were serotyped and tested for the presence of specific virulence genes. A completely randomized design with a 6 × 3 × 15 factorial arrangement was utilized to analyze the results for Salmonella prevalence between processing plants. Means were separated using Fishers protected least significant difference when P ≤ 0.05. For Salmonella prevalence between processing plants, differences (P < 0.0001) were observed in the 6 plants tested where the maximum and minimum prevalence was 29.6 and 7.4%, respectively. As expected, there was a difference (P < 0.0001) in the recovery of Salmonella because of sampling time. Salmonella prevalence at time A (36%) was significantly higher, whereas there was no difference between time B (12%) and C (9%). There was a location effect (P < 0.0001) for the prevalence of Salmonella with the head puller, picker, cropper, and scalder having a significantly higher prevalence when compared with several other locations. At sampling time C, a trend toward a difference (P = 0.0899) was observed for Salmonella prevalence between the 6 plants, whereas significant differences were observed because of location (P = 0.0031). Five prominent Salmonella enterica serovars were identified, including Kentucky, Schwarzengrund, Enteritidis, Liverpool, and Typhimurium with S. Kentucky being the most prevalent. PCR analysis of 8 Salmonella virulence genes showed that the invA, sipB, spiA, sseC, and fimA were detected in all isolates, whereas genes carried on plasmids and/or fimbriae varied remarkably among all isolates. This study established Salmonella prevalence and persistence in poultry processing facilities after antimicrobial application through sanitization procedures which could result in contamination of poultry carcasses and food safety risks because of poultry meat.
Key words: Salmonella, prevalence, sanitization, poultry processing, virulence
Introduction
Poultry integrators in the United States are continually working toward producing safe poultry meat (NCC, 2020). To achieve this goal, it is critical for integrators to follow regulatory programs and procedures that are directed at controlling foodborne pathogens during broiler processing (Simmons et al., 2003; McKee, 2012; NCC, 2020). Foodborne infections remain a public health challenge in the United States. In 2018, the Foodborne Diseases Active Surveillance Network (FoodNet) of the CDC identified over 25,000 infections and approximately 6,000 hospitalizations because of foodborne pathogens (CDC, 2018). Salmonella infections was reported as the second most common foodborne infection accounting for 9,084 infections at a rate of 18.3 cases per 100,000 people (Tack et al., 2019). Additionally, Salmonella infections caused the most hospitalizations and deaths in the same year with the top 3 Salmonella serotypes causing salmonellosis being Enteritidis, Newport, and Typhimurium. The Food Safety and Inspection Service (FSIS) of the United States Department of Agriculture (USDA) also identified Salmonella in 3.7% of all the chicken carcasses sampled under the Hazard Analysis Critical Control Point verification program in 2014, and the top 2 serotypes identified were Kentucky and Enteritidis (USDA-FSIS, 2016). Salmonella control during the processing of poultry meat is crucial. Among all food commodities, chicken causes the most Salmonella outbreak-associated illnesses, hospitalizations, and deaths than any other food (CDC, 2016). Moreover, per capita consumption of broiler meat has continued to increase for the past 2 decades and is expected to continue to increase in the future (NCC, 2019).
Efficient cleaning and sanitization procedures are part of the plant operating procedures that are intended to control foodborne pathogens, including Salmonella and Campylobacter in poultry processing facilities (Olsen et al., 2003; Potter et al., 2012; Lebron, 2013). Poultry processing facilities clean and sanitize their plant and equipment after processing meat to produce safe, wholesome products and to eliminate any pathogenic microorganisms that may be present. However, some microorganisms are able to adhere to food processing equipment surfaces and remain active after cleaning and sanitization (Chmielewski and Frank, 2003; Carpentier, 2011; Fagerlund et al., 2017). The persistence of such pathogenic microorganisms on the equipment surface could result in cross-contamination of a pathogen-free flock during processing (Rasschaert et al., 2008). Antimicrobial application or sanitization is a step conducted after the cleaning process. Sanitizers such as sodium hypochlorite, quaternary ammonium compounds, and hydrogen peroxides are among the antimicrobial agents approved by the USDA for the disinfection of poultry carcasses as well as for the sanitization of poultry processing equipment and inside the facility (USDA-FSIS, 2017a). The potency of these sanitizing agents is essential for microbial inactivation during the cleaning and sanitization procedure. The efficacy of different antimicrobial agents at inactivating foodborne pathogens like Salmonella during poultry processing and on retail poultry meat has been established (Firildak et al., 2015; Kim et al., 2017; Li et al., 2017; Moore et al., 2017; Zhang et al., 2019). Others have reported the ability of sanitizers like chlorine to reduce foodborne pathogens on different food-contact surfaces encountered in food processing plants (Shen et al., 2012; Schlisselberg and Yaron, 2013; Smith et al., 2015).
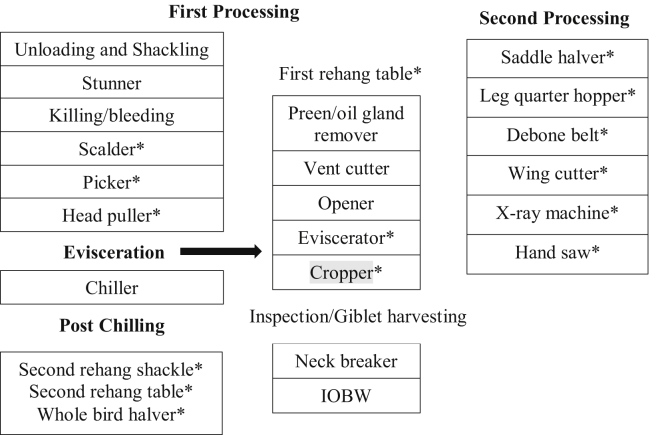
Recovery of Salmonella from poultry transport equipment, the slaughterhouse, and the processing environment has been documented, but minimal information is available about its recovery from poultry processing equipment after antimicrobial application (Simmons et al., 2003; Reiter et al., 2007; Lestari et al., 2009; Mezal et al., 2014; Khan et al., 2018). A study conducted in France recovered Campylobacter jejuni from the picker and eviscerator before cleaning and disinfection, and this pathogen persisted on the equipment and was recovered again after the disinfection procedure (Peyrat et al., 2008). Similarly, several Salmonella serotypes were recovered from processing equipment of Malaysian wet markets and a small-scale processing plant (Nidaullah et al., 2017). The persistence and prevalence of Salmonella on poultry processing equipment highlighted in Figure 1, not only after poultry meat processing but also after cleaning and sanitizing the processing equipment has not been well documented in the United States. The equipment could be indirectly contaminated with pathogens that reside in the gut of live birds and cross-contaminate poultry meat if improperly cleaned. Hence, it is imperative to understand the extent to which Salmonella persist in the poultry processing environment, as this knowledge could improve the effectiveness of current Salmonella control measures. In the current study, the prevalence of Salmonella in the poultry processing environment and its persistence on processing equipment after the cleaning and sanitization procedures within different poultry processing plants is reported. The recovered isolates were further identified by serotyping and characterized based on virulence genes.
Figure 1.
Poultry processing equipment layout with ∗sampling points. Abbreviation: IOBW, inside/outside bird washer.
Materials and methods
Experimental Design
Six different poultry processing plants (designated 1, 2, 3, 4, 5, and 6) belonging to 3 poultry integrators in the southern region of the United States were visited from January 2018 to January 2019. Each plant has a shift dedicated to thorough cleaning and sanitization, and each were visited for sample collection on 3 different days. Samples were collected from 15 different pieces of equipment (locations) at 3 different time periods described as A–after processing, B–after cleaning, and C–after sanitization. A total of 135 samples were collected at each plant, making 45 samples per time period. The processing equipment sampled are highlighted in Figure 1.
Sample Collection
Swab samples of poultry processing equipment were collected using 3M sponge-stick with 10 mL buffered peptone water or neutralizing broth (SSL10NB, 3M Co. St. Paul, MN). The surface of each piece of equipment from Figure 1 was sampled 3 times. Each piece of equipment was swabbed horizontally by covering a surface area of about 13 cm2 for 30 s, and the same spot was swabbed 3 times as described above. Each sample was immediately placed in a cooler with ice packs. All samples were immediately transported in a cooler containing ice and analyzed immediately upon reaching the lab at Mississippi State University.
Identification of Isolates
Media was purchased from Fisher Scientific, Hampton, NH, unless otherwise specified. Detection and isolation of Salmonella from samples was carried out following the USDA MLG 4.09 conventional method (USDA-FSIS, 2017b). Each sample was pre-enriched in 50 mL of buffered peptone water (BPW, BD218105) and incubated at 37°C for 20 to 24 h. After pre-enrichment, an aliquot of 100 μL and 500 μL was transferred into 10 mL of modified rappaport vassiliadis (mRV, CM0910 B) and tetrathionate broth (TT Hajna, BD249120) respectively and incubated at 42°C for 22 to 24 h. One loop full of culture in mRV and TT was subsequently streaked, in duplicate, onto brilliant green sulfa (BGS, BD271710) and xylose lysine tergitol 4 (XLT4, BD223420) agar. The agar plates were incubated at 37°C for 18 to 24 h. Afterward, presumptive Salmonella colonies were selected for biochemical testing by inoculating into triple sugar iron (BD226540) and lysine iron agar (BD211363) slants in tandem. The positive Salmonella isolates that were recovered from the slants at time period C (post sanitization) were sent to the National Veterinary Service Laboratory (NVSL, Ames, IA) for serotyping and used for all subsequent analysis.
Isolate Collection and Preservation
Following identification of the isolates recovered after sanitization, a loop of culture from Salmonella-positive triple sugar iron slant was streaked onto XLT4 plates and incubated at 37°C for 24 h. A distinct Salmonella colony was picked from the XLT4 plate and streaked onto tryptic soy agar (BD236950) and incubated. Afterward, cells were harvested and preserved in cryotubes containing a 20% glycerol solution (G33500, Fisher Scientific) and stored at −80°C.
DNA Extraction
For extraction of Salmonella DNA, isolates recovered after sanitization were streaked from the cryotubes onto tryptic soy agar plates. Following incubation, colonies from each plate were inoculated into 10 mL tryptic soy broth (BD211825) and incubated at 37°C for 24 h. After incubation, 2 mL of the culture was transferred to a microcentrifuge tube and centrifuged at 5,000 × g for 3 min followed by removing the supernatant from the pellet. This process was continued until all 10 mL of the culture was centrifuged. The pellets were resuspended twice in 2 mL phosphate buffered saline (J75889AE). The DNA was extracted from the pellet using the QIAamp DNA mini kit (51,304, Qiagen, Hilden, Germany) and analyzed for quantity and purity using a Nanodrop one UV-VIS spectrophotometer (Thermo Fisher Scientific, Waltham, MA) with a 260/280 nm ratio of 1.8 to 2.0 purity standard.
PCR Detection of Salmonella Virulence-Related Genes
Eight genes attributed to virulence in Salmonella were selected. Four of the genes (invA, sipA, spiA, and sseC) are located within the Salmonella pathogenicity islands (SPI) 1 and 2, 3 targets (spvB, spvC, and pefA) are found on the Salmonella virulence plasmid (pSLT), and 1 gene (fimA) encodes Type 1 fimbriae of Salmonella (Fàbrega and Vila, 2013; Suez et al., 2013).
After DNA extraction from each isolate, the presence of the virulence genes was determined using a PCR technique previously described by Oliveira et al. (2002). DNA from reference strains of S. Typhimurium ATCC 14028, S. Enteritidis ATCC 4931, and S. Heidelberg ATCC 8326 were included. All primers that were used to detect virulence genes are listed in Table 1. For each virulence gene, amplification took place in a 25 μL reaction containing 12.5 μL 2× Promega GoTaq master mix (M7122, Promega, Madison, WI), 0.5 μL each (10 μmol) F/R primers, 10.5 μL nuclease-free water, and 1 μL (80–120 ng) of DNA template. PCR was carried out in a Eppendorf EP gradient master cycler (Eppendorf Biotech Company, Hamburg, Germany) using the following cycling conditions: Initial denaturation at 98°C for 3 min, followed by 40 cycles of denaturation at 98°C for 30 s, annealing at 55°C for 30 s, and extension at 72°C for 30 s, with a final extension at 72°C for 5 min. Gel electrophoresis of PCR products was carried out on 2% agarose gel (A201100, GoldBio, St. Louis, MO) containing SYBR safe DNA gel stain (Invitrogen S33102, Fisher Scientific) for visualization and a 100 bp DNA ladder (D001500, GoldBio) as a marker.
Table 1.
List of primers used in this study.
| Target gene | Primers | Sequence | Product size | Reference |
|---|---|---|---|---|
| invA | invA-F | GTGAAATTATCGCCACGTTCGGGCAA | 284 | Rahn et al., 1992 |
| invA-R | TCATCGCACCGTCAAAGGAACC | |||
| sipB | sipB-F | GGACGCCGCCCGGGAAAAACTCTC | 875 | Skyberg et al., 2006 |
| sipB-R | ACACTCCCGTCGCCGCCTTCACAA | |||
| spiA | spiA-F | CCAGGGGTCGTTAGTGTATTGCGTGAGATG | 550 | Skyberg et al., 2006 |
| spiA-R | CGCGTAACAAAGAACCCGTAGTGATGGATT | |||
| sseC | sseC-F | ATGAATCGAATTCACAGTAA | 1,455 | Bhowmick et al., 2011 |
| sseC-R | TTAAGCGCGATAGCCAGCTA | |||
| spvB | spvB-F | CTATCAGCCCCGCACGGAGAGCAGTTTTTA | 717 | Tarabees et al., 2017 |
| spvB-R | GGAGGAGGCGGTGGCGGTGGCATCATA | |||
| spvC | spvC-F | ACTCCTTGCACAACCAAATGCGGA | 571 | Chaudhary et al., 2015 |
| spvC-R | TGTCTTCTGCATTTCGCCACCATCA | |||
| fimA | fimA-F | CCTTTCTCCATCGTCCTGAA | 85 | Naravaneni and Jamil, 2005 |
| fimA-R | TGGTGTTATCTGCCTGACCA | |||
| pefA | pefA-F | GCGCCGCTCAGCCGAACCAG | 157 | Tarabees et al., 2017 |
| pefA-R | GCAGCAGAAGCCCAGGAAACAGTG |
Statistical Analysis
Differences in Salmonella prevalence on equipment amongst plants over time were determined by analysis of variance in the General Linear Model using SAS software v 9.4 (SAS Institute, 2013; Steel and Torrie, 1980). A 6 (plants) × 3 (time period) × 15 (locations) factorial arrangement of treatments in a completely randomized design was used to determine the effect of plant, time, and location on Salmonella prevalence. When significant differences (P ≤ 0.05) were observed, Fisher's protected Least Significant Difference Test (P ≤ 0.05) was used to separate the means.
Results
Prevalence of Salmonella on Poultry Processing Equipment Because of Plants
Prevalence of Salmonella on poultry processing equipment among the 6 poultry processing plants based on overall sampling times was compared (Table 2). Overall, plants 1, 2, and 4 from integrators 1 and 2 had higher prevalence (P < 0.001) of Salmonella on their processing equipment when compared with plants 5 and 6 from integrator 3. When looking at prevalence within an integrator, there was no significant difference (P = 0.22) in Salmonella prevalence on equipment between plants 1 and 2 within integrator 1. Similarly, no differences were observed (P = 0.76) in prevalence between plants 5 and 6 within integrator 3. In contrast, Salmonella prevalence differed (P = 0.03) on the processing equipment of plants 3 and 4 within integrator 2.
Table 2.
Overall prevalence of Salmonella on poultry processing equipment of different processing plants.
| Integrator | Plant | Prevalence (%) |
|---|---|---|
| 1 | 1 | 29.6a |
| 2 | 23.7a,b | |
| 2 | 3 | 17.8b,c |
| 4 | 28.2a | |
| 3 | 5 | 7.4d |
| 6 | 8.9c,d |
a-dEach plant was visited 3 times. Means with different superscripts indicate significant differences.
N = 135, SEM = 3.41, P < 0.001.
Prevalence of Salmonella on Poultry Processing Equipment Because of Time
Salmonella prevalence was higher (P < 0.001) on the equipment at the end of the day's processing (time period A, 36%) compared with after the cleaning procedure, which included scrubbing and washing with detergent (time period B, 12%). In contrast, there were no differences (P = 0.39) in prevalence between time periods B and C (9%), which was after sanitization (Table 3).
Table 3.
Prevalence of Salmonella on poultry processing equipment of different processing plants over 3 time periods.
| Time | Prevalence (%) |
|---|---|
| A—Postprocessing | 36.3a |
| B—Postcleaning | 12.2b |
| C—Postsanitization | 9.3b |
a,bMeans with different superscripts indicate significant differences.
N = 270, SEM = 2.41, P < 0.001.
Prevalence of Salmonella on Poultry Processing Equipment Because of Location
Salmonella prevalence differed (P < 0.0001) among the equipment (locations) that was sampled in this study (Table 4). Salmonella was observed to persist more on some of the first processing equipment on the kill line, including the head puller, picker, and scalder when compared with others on the evisceration line, like the eviscerator in all the processing plants. Prevalence was similar (P > 0.05) on most of the second processing equipment among all plants.
Table 4.
Prevalence of Salmonella on poultry processing equipment of different processing plants.
| Processing step | Location1 | Prevalence (%) |
|---|---|---|
| First processing (kill line) | Head puller | 40.7a |
| Picker | 53.7a | |
| Scalder | 42.6a | |
| First rehang | First rehang | 20.4b,c |
| First processing (evis line) | Cropper | 44.4a |
| Eviscerator | 24.1b | |
| Second rehang | Second rehang shackle | 7.4c,d |
| Second rehang table | 0d | |
| Second processing | Debone belt | 13.0b,c,d |
| Hand saw | 3.7d | |
| Halver | 9.3b,c,d | |
| Leg quarter hopper | 9.3b,c,d | |
| Saddle halver | 5.6c,d | |
| Wing cutter | 9.3b,c,d | |
| X-ray belt | 5.6c,d |
a-dSuperscripts indicate significant differences. N = 54, SEM = 5.39, P = 0.001.
Equipment within each processing plants sampled. Means with different.
Persistence of Salmonella on Processing Equipment After Sanitization Because of Plant and Location
After antimicrobial application (sanitization), there was a trend (P = 0.09) where less Salmonella could be recovered from processing equipment (Table 5). For persistence of Salmonella on the equipment because of plants, there were no significant differences in prevalence between plants 1 and 2 (P = 0.76), which are managed by integrator 1, plants 3 and 4 (P = 0.76) managed by integrator 2, and plant 5 and 6 (P = 0.76) managed by integrator 3. However, plants 3 and 4 had the highest percent prevalence compared to the other plants. Integrator 3 had the lowest percent prevalence when compared with integrators 1 and 2. Moreover, Salmonella was recovered from all the processing plants after sanitization except plant 6 where there was no Salmonella found on the equipment. When looking at the persistence of Salmonella on the equipment (location) after sanitization, recovery of Salmonella was observed to be significantly higher (P = 0.0002) on first processing equipment, including the cropper, scalder, picker, and head puller when compared with several other types of equipment (Table 6). There were no differences (P = 0.62) in prevalence between the cropper and scalder, picker, and head puller. However, significant differences (P = 0.05) were observed when the cropper was compared with the debone belt and leg quarter hopper, which belong to the second processing equipment and were the only second processing equipment where Salmonella persisted.
Table 5.
Recovery of Salmonella from poultry processing equipment of different processing plants after sanitization.
| Integrator | Plant | Prevalence (%) |
|---|---|---|
| 1 | 1 | 8.9 |
| 2 | 11.1 | |
| 2 | 3 | 15.6 |
| 4 | 17.8 | |
| 3 | 5 | 2.2 |
| 6 | 0.0 |
N = 45, SEM = 5.02, P = 0.09.
Table 6.
Recovery of Salmonella from poultry processing equipment of different processing plants after sanitization.
| Processing step | Location1 | Prevalence (%) |
|---|---|---|
| First processing (kill line) | Head puller | 27.8a |
| Picker | 27.8a | |
| Scalder | 27.8a | |
| First rehang | First rehang | 0.0b |
| First processing (evis line) | Cropper | 33.3a |
| Eviscerator | 0.0b | |
| Second rehang | Second rehang shackle | 0.0b |
| Second rehang table | 0.0b | |
| Second processing | Debone belt | 11.1a,b |
| Hand saw | 0.0b | |
| Halver | 0.0b | |
| Leg quarter hopper | 11.1a,b | |
| Saddle halver | 0.0b | |
| Wing cutter | 0.0b | |
| X-ray belt | 0.0b |
a,bSuperscripts indicate significant differences. N = 18, SEM = 7.93, P = 0.0002.
Equipment within each processing plants sampled. Means with different.
Serotypes of Isolates Recovered From Processing Equipment After Sanitization
A total of 25 Salmonella isolates were recovered from various pieces of equipment from different processing plants after the sanitization (time period C) procedure. The various Salmonella serovars isolated are listed in Table 7. The recovered isolates belonged to 5 distinct Salmonella serovars. The most prevalent serovar isolated was Salmonella Kentucky (n = 12, 48%) followed by Salmonella Schwarzengrund (n = 5, 20%). Four isolates were identified as Salmonella Enteritidis (16%), 3 were Liverpool (12%), and only 1 (4%) was Salmonella Typhimurium.
Table 7.
Salmonella serovars recovered after sanitization of different processing plants.
| Salmonella enterica | No. of isolates | Prevalence (%) |
|---|---|---|
| Salmonella Enteritidis | 4 | 16 |
| Salmonella Typhimurium | 1 | 4 |
| Salmonella Schwarzengrund | 5 | 20 |
| Salmonella Kentucky | 12 | 48 |
| Salmonella Liverpool | 3 | 12 |
| Total | 25 |
Virulence of Salmonella Isolates Recovered From Processing Equipment After Sanitization
All the isolates that were recovered after sanitization were further characterized by examining 8 virulence genes in Salmonella. At least 4 virulence genes (invA, sipB, spiA, and sseC) found within SPI 1 and 2, and 1 related to fimbriae (fimA) were detected in all 25 Salmonella isolates (Table 8). Detection of the genes located on Salmonella virulence plasmid (pSLT) varied among the isolates. Among the genes carried on the plasmid, spvB (23/25) and pefA (24/25) were detected more frequently from the isolates spvC (9/25) was detected less frequently (Table 8).
Table 8.
Salmonella virulence genes detected in the isolates.
| Virulence genes | Location | No. of isolates positive (%) | No of isolates negative (%) |
|---|---|---|---|
| invA | SPI-1 | 25 (100%) | 0 |
| sipB | 25 (100%) | 0 | |
| spiA | SPI-2 | 25 (100%) | 0 |
| sseC | 25 (100%) | 0 | |
| spvB | pSLT plasmid | 23 (92%) | 2 (8%) |
| spvC | 9 (36%) | 16 (64%) | |
| pefA | 24 (96%) | 1 (4%) | |
| fimA | fimbriae | 25 (100%) | 0 |
Discussion
Overall Prevalence of Salmonella by Plants, Time Periods, and Location
For several years, non-typhoidal Salmonella has remained a pathogen of importance to public health because it causes gastroenteritis in both developed and developing countries. Majowicz et al. (2010) estimated the global burden of Salmonella infection, gastroenteritis that were foodborne to be 80.3 million cases each year. According to the CDC, consumption of contaminated chicken meat is still a significant source of Salmonella infection in the United States (CDC, 2016). In other parts of the world where chicken meat is readily available, Salmonella contamination has been reported to cause illnesses and deaths (Barua et al., 2014; Nidaullah et al., 2017). Contamination can occur during different stages of poultry processing, including through the improper cleaning of processing equipment (Olsen et al., 2003; Lestari et al., 2009). Although Salmonella prevalence in raw chicken meat has been reported, this study presents the persistence of Salmonella on processing equipment after chickens have been processed through the cleaning and sanitization process.
In the present study, the presence of Salmonella on processing equipment of different poultry processing plants was tracked, and Salmonella was recovered from all the processing plants sampled. Many factors may have contributed to the recovery of Salmonella from the plants that were sampled. These include the antimicrobial that was used, the cleaning procedure, and the attachment of Salmonella to the equipment as biofilms. It has been previously suggested that Salmonella could persist in an environment by attaching firmly to abiotic surfaces, thus rendering antimicrobial applications ineffective (Gram et al., 2007). Although prevalence was lower in some plants compared with others, the pathogen was recovered from all the plants immediately after processing. It is expected that there would be a high prevalence of Salmonella on the equipment after processing chickens (time period A) because Salmonella is a commensal pathogen of the gut for many animals including poultry (Cosby et al., 2015). There was a reduction in the recovery of Salmonella from time periods A to B (after cleaning), but no significant reduction was observed in prevalence between time periods B and C (after sanitization). The process of cleaning with detergent, some of which may have low levels of antimicrobial activity, could explain the reduction in bacterial contamination on the equipment surfaces. The cleaning process in all the plants visited required both physical and chemical activity. This process involved physical scrubbing of the equipment with a sponge, brush, and chemical detergent. Moreover, the application of sanitizers like chlorine or a quaternary ammonium compound on the equipment is presumed to further reduce bacterial contamination. It is possible that Salmonella have persisted on the equipment surface over time and thus are able to tolerate the antimicrobials that are used for sanitization. Previous studies have reported that Salmonella could acquire tolerance to antimicrobials when exposed to subinhibitory concentrations over time (Condell et al., 2012; Obe et al., 2018). Furthermore, high prevalence of Salmonella was observed for some of the first processing equipment, whereas there was no change in prevalence for several pieces of second processing equipment. The first processing of birds including evisceration, where the eviscerator removes the internal content of the carcass, which can cause contamination of both the carcass and the equipment with Salmonella (Russell and Walker, 1997). However, most processing plants use equipment spray with antimicrobial agents to reduce contamination during poultry processing (Bourassa, 2018).
Prevalence of Salmonella After Sanitization (Time Period C) by Plants and Location
To determine whether Salmonella persists on the equipment after sanitization, prevalence at time period C was only analyzed. Salmonella prevalence was considerably higher in plants managed by integrators 1 and 2 compared with integrator 3, which has at least 1 plant where there was no Salmonella recovered from its equipment after sanitization. Similar to overall prevalence, plants with higher overall prevalence had higher prevalence after sanitization, which suggests that the antimicrobials that were used may be ineffective, or the cleaning procedure employed by the plant may not be adequate to reduce microbial contamination on the equipment surfaces. Another possible explanation is that the plants with persistent Salmonella on the equipment surface had a higher initial bacterial load as seen in overall prevalence at sampling time period A (after processing). Furthermore, Salmonella was found to persist more on first processing equipment than several other pieces of equipment. Some of the equipment are located on the kill side of the processing plant and could easily be contaminated with Salmonella from a positive flock and therefore require more rigorous cleaning. Studies have reported prevalence of Salmonella and Campylobacter on equipment like scalders, pickers, and eviscerators at the end of processing. However, similar to our findings, Campylobacter has been recovered from the picker, eviscerator, and conveyor belt before and after antimicrobial application, whereaas Salmonella was found to be prevalent on the picker after sanitization (Trampel, 2000; Olsen et al., 2003; Peyrat et al., 2008). Other equipment, especially those used in the second processing area, had significantly lower prevalence after processing, and no bacteria was recovered from the equipment after sanitization. This may be due to a lower initial bacterial load coupled with different interventions aimed at reducing Salmonella contamination during poultry processing. Antimicrobial intervention in the chiller is 1 of the critical control measures against Salmonella during poultry processing, but recovery has been reported from carcasses at the exit chiller (Parveen et al., 2007; Wideman et al., 2016). This could cause contamination of second processing equipment that have been thoroughly cleaned and sanitized. Also, of all the equipment sampled, the head puller was positive for Salmonella at all the 6 plants. Similarly, debone belt and leg quarter hopper were positive after sanitization. A possible explanation is that the equipment may be hard to reach for thorough cleaning and may therefore require more attention during the cleaning procedure.
Serotypes of Salmonella Isolates Recovered After Sanitization
There were 5 distinct Salmonella serovars identified in this study including Kentucky, Schwarzengrund, Enteritidis, Liverpool, and Typhimurium. S. Enteritidis and S. Typhimurium are 2 of the top 3 serovars identified in salmonellosis. According to CDC data, the number of infections caused by S. Enteritidis has increased from 2008 to 2018 and that the source of the infections could be traced to poultry and eggs (CDC, 2018). While S. Enteritidis was the most common serotype identified by USDA-FSIS from poultry establishments over a 3-year period, recovery of S. Typhimurium was reduced in the same establishments over the same 3-year period (CDC, 2018). Salmonella Kentucky is the most prevalent serovar identified in this study, and it has emerged as the top serovar identified in live poultry, turkey, and chicken meat (Lestari et al., 2009; Foley et al., 2011). Furthermore, the USDA data showed S. Kentucky was the most prevalent serovar from routine testing of chicken samples as part of the Hazard Analysis Critical Control Point verification program (USDA-FSIS, 2016). Salmonella Schwarzengrund was the second most prevalent serovar that was isolated from processing equipment. This serovar is not among the top 10 serovars commonly identified in poultry meat by CDC or USDA but has been implicated in multistate foodborne outbreaks that resulted in illnesses and hospitalizations (CDC, 2007). In addition, S. Kentucky and S. Schwarzengrund that have been previously recovered from poultry products have expressed resistance to multiple antibiotics of clinical importance (Aarestrup et al., 2007; Lestari et al., 2009). The reduction in the recovery of Typhimurium from poultry and infection in humans has been linked to vaccination and better production practices (Dórea et al., 2010). Vaccinating commercial poultry against Typhimurium and Enteritidis could help reduce the incidence of salmonellosis that is caused by these serovars, but also increase infections caused by other serovars like Kentucky and Schwarzengrund to which vaccines have not been developed (Foley et al., 2011). Therefore, vaccinating poultry against emerging strains of Salmonella implicated in salmonellosis may help to further control Salmonella contamination in poultry meat.
Virulence of Salmonella Isolates Recovered After Sanitization
The ability of Salmonella to cause infection in humans has been extensively studied using S. Typhimurium. In this study, to determine the extent to which the recovered isolates could cause infection, the presence of virulence genes implicated in colonization by S. Typhimurium were examined. These genes are located within the Salmonella pathogenicity islands (SPI 1 & 2), virulence plasmid (pSLT), and the fimbrial subunit. Their functions include host recognition and invasion, survival, and replication within the epithelial cells, inhibition of inflammatory response and actin polymerization, and adhesion to specific epithelial cells (Fàbrega and Vila, 2013; Mezal et al., 2014). Many of the genes tested with the exception of the 1 found in the plasmid were detected in the recovered isolates. This observation is in agreement with other studies, where similar genes associated with multiple Salmonella strains that were isolated from poultry houses, chicken samples, and clinical samples were compared, with similarities found in their virulence. In fact, the poultry and clinical isolates shared virulence genotypes, which suggests that the poultry isolates can cause infection in humans (Diarra et al., 2014; Mezal et al., 2014; Yang et al., 2016; Rauch et al., 2018). Similarly, the findings in this study suggest that if the recovered isolates were to contaminate chicken meat and safe food handling practices were not followed, salmonellosis could occur. Notably, all the recovered S. Kentucky carried at least one virulence plasmid gene, even though S. Kentucky is not the most reported serovar in Salmonella infection. Salmonella virulence plasmids have been suggested to play a pivotal role in Salmonella infection (Guiney et al., 1995; Yang et al., 2016). Barua et al. (2014) found similarities in the PFGE profile of S. Kentucky from poultry and human sources. Likewise, Rauch et al. (2018) observed the same Salmonella Kentucky sequence types in isolates from chicken meat and clinical samples. Additionally, studies have found that Salmonella isolates carrying the virulence plasmid possess resistance to multiple antibiotics, which could make treatment with clinically important antibiotics challenging (Barua et al., 2014; Yang et al., 2016).
Conclusion
In conclusion, contaminated processing equipment could serve as a potential source of cross-contamination of poultry carcasses during poultry processing because pathogens are able to survive the cleaning and sanitization procedure, thus causing food safety risks (Peyrat et al., 2008; Perez-Arnedo and Gonzalez-Fandos, 2019). It is critical to mention that all the processing plants that were visited in this study dedicated substantial time to the cleaning and sanitization procedure of the equipment and facility between the end of the shift and the next processing, but more effort may be required to address Salmonella contamination during poultry processing. The prevalence of S. Kentucky observed in this study is worth further exploration because virulence genes previously identified in S. Typhimurium and S. Enteritidis infection were detected. Also, further examination of antibiotic resistance profiles of the recovered isolates would be noteworthy.
A drawback observed in this study is the lack of data on the prevalence of Salmonella in the flocks processed at each of the plant sampled. This information could help to link the serovars of Salmonella recovered at each plant to the flock processed and reveal whether the serovars have been persisting on the equipment from a previous flock or processing day. The conclusions in this study could also be better supported by tracking prevalence of Salmonella on the equipment to the chickens processed by the plant at retail level. Regardless, this study fills some gaps in knowledge regarding the efficiency of the cleaning and sanitization procedure to reduce Salmonella contamination. This information could further be utilized to determine the mechanism by which the recovered isolates persist in the processing environment.
Acknowledgments
This publication is a contribution of the Mississippi Agricultural and Forestry Experiment Station. This material is based upon work that is supported by the U. S. Department of Agriculture, Hatch projects under accession numbers of MIS-322340.
Conflict of Interest Statement: There are no conflicts of interest to disclose.
References
- Aarestrup F.M., Hendriksen R.S., Lockett J., Gay K., Teates K., McDermott P.F., White D.G., Hasman H., Sørensen G., Bangtrakulnonth A., Pornreongwong S., Pulsikarn C., Angulo F.J., Garner-Smidt P. International spread of multidrug-resistant Salmonella Schwarzengrund in food products. Emerg. Infect. Dis. 2007;13:726–731. doi: 10.3201/eid1305.061489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barua H., Biswas P.K., Talukder K.A., Olsen K.E.P., Christensen J.P. Poultry as a possible source of non-typhoidal Salmonella enterica serovars in humans in Bangladesh. Vet. Microbiol. 2014;168:372–380. doi: 10.1016/j.vetmic.2013.11.020. [DOI] [PubMed] [Google Scholar]
- Bhowmick P.P., Devegowda D., Darshanee Ruwandeepika H.A., Karunasagar I., Karunasagar I. Presence of Salmonella pathogenicity island 2 genes in seafood-associated Salmonella serovars and the role of the sseC gene in survival of Salmonella enterica serovar Weltevreden in epithelial cells. Microbiol. 2011;157:160–168. doi: 10.1099/mic.0.043596-0. [DOI] [PubMed] [Google Scholar]
- Bourassa D.V. Antimicrobial use in poultry processing. Food Saf. Mag. 2018 Accessed June 2019. https://www.foodsafetymagazine.com/magazine-archive1/december-2017january-2018/antimicrobial-use-in-poultry-processing/ [Google Scholar]
- Carpentier B. 2011. Biofilms and Microorganisms on Surfaces after Cleaning and Disinfection. Accessed May 2020. https://www.foodsafetymagazine.com/magazine-archive1/aprilmay-2011/biofilms-and-microorganisms-on-surfaces-after-cleaning-and-disinfection/ [Google Scholar]
- Center for Disease Control and Prevention (CDC). 2007. Salmonella Schwarzengrund Outbreak Investigation, August 2007. Accessed June 2019. https://www.cdc.gov/salmonella/schwarzengrund.html. [Google Scholar]
- Center for Disease Control and Prevention (CDC). Annual report.; 2016. Surveillance for Foodborne Disease Outbreaks United States, 2016. Accessed June 2019. https://www.cdc.gov/fdoss/pdf/2016_foodborneoutbreaks_508.pdf?deliveryname=dm9453. [Google Scholar]
- Center for Disease Control and Prevention (CDC). 2018. FoodNet 2018 preliminary data. Accessed June 2019. https://www.cdc.gov/foodnet/reports/prelim-data-intro-2018.html. [Google Scholar]
- Chaudhary J.H., Nayak J.B., Brahmbhatt M.N., Makwana P.P. Virulence genes detection of Salmonella serovars isolated from pork and slaughterhouse environment in Ahmedabad, Gujarat. Vet. World. 2015;8:121–124. doi: 10.14202/vetworld.2015.121-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chmielewski R.A.N., Frank J.F. Biofilm formation and control in food processing facilities. Compr. Rev. Food Sci. Food Saf. 2003;2:22–32. doi: 10.1111/j.1541-4337.2003.tb00012.x. [DOI] [PubMed] [Google Scholar]
- Condell O., Iversen C., Cooney S., Power K.A., Walsh C., Burgess C., Fanning S. Efficacy of Biocides used in the modern food Industry to control Salmonella enterica, and links between Biocide tolerance and resistance to clinically Relevant antimicrobial compounds. Appl. Environ. Microbiol. 2012;78:3087–3097. doi: 10.1128/AEM.07534-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosby D.E., Cox N.A., Harrison M.A., Wilson J.L., Buhr R.J., Fedorka-Cray P.J. Salmonella and antimicrobial resistance in broilers: a review. J. Appl. Poult. Res. 2015;24:408–426. [Google Scholar]
- Diarra M.S., Delaquis P., Rempel H., Bach S., Harlton C., Aslam M., Pritchard J., Topp E. Antibiotic resistance and diversity of Salmonella enterica serovars associated with broiler chickens. J. Food Prot. 2014;77:10–49. doi: 10.4315/0362-028.JFP-13-251. [DOI] [PubMed] [Google Scholar]
- Dórea F.C., Cole D.J., Hofacre C., Zamperini K., Mathis D., Doyle M.P., Lee M.D., Maurer J.J. Effect of Salmonella vaccination of breeder chickens on contamination of broiler chicken carcasses in integrated poultry operations. Appl. Environ. Microbiol. 2010;76:7820–7825. doi: 10.1128/AEM.01320-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fàbrega A., Vila J. Salmonella enterica serovar Typhimurium skills to succeed in the host: virulence and regulation. Clin. Microbiol. Rev. 2013;26:308–341. doi: 10.1128/CMR.00066-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagerlund A., Moretro T., Heir E., Briandet R., Langsrud S. Cleaning and disinfection of biofilms composed of Listeria monocytogenes and background microbiota from meat processing surfaces. Appl. Environ. Microbiol. 2017;83:e010146-17. doi: 10.1128/AEM.01046-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firildak G., Asan A., Goren E. Chicken carcasses bacterial concentration at poultry slaughter facilities. Asian J. Biol. Sci. 2015;8:16–29. [Google Scholar]
- Foley S., Nayak R., Hanning I.B., Johnson T.J., Han J., Rickey S.C. Population dynamic of Salmonella enterica serotypes in commercial eggs and poultry production. Appl. Environ. Microbiol. 2011;77:4273–4279. doi: 10.1128/AEM.00598-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gram L., Bagge-Ravn D., Ng Y.Y., Gymoese P., Vogel B.F. Influence of food soiling matrix on cleaning and disinfection efficiency on surface attached Listeria monocytogenes. Food Control. 2007;18:1165–1171. [Google Scholar]
- Guiney D.G., Fang F.C., Krause M., Libby S., Buchmeier N.A., Fierer J. Biology and clinical significance of virulence plasmids in Salmonella serovars. Clin. Infect Dis. 1995;21(Suppl. 2):S146–S151. doi: 10.1093/clinids/21.supplement_2.s146. [DOI] [PubMed] [Google Scholar]
- Khan A.S., Georges K., Rahamann S., Abdela W., Adesiyun A.A. Prevalence and serotypes of Salmonella spp. on chickens sold at retail outlets in Trinidad. PLoS One. 2018;13:e0202108. doi: 10.1371/journal.pone.0202108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.A., Park S.H., Lee S.I., Owens C.M., Ricke S.C. Assessment of chicken carcass microbiome responses during processing in the presence of commercial antimicrobials using a next generation sequencing approach. Sci. Rep. 2017;7:43354. doi: 10.1038/srep43354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebron J. Plant Engineering.; 2013. Sanitation Standard Operating Procedure in the Meat and Poultry Food Manufacturing Plant. Accessed June 2019. https://www.plantengineering.com/articles/sanitation-standard-operating-procedure-in-the-meat-and-poultry-food-manufacturing-plant/ [Google Scholar]
- Lestari S.I., Han F., Wang F., Ge B. Prevalence and antimicrobial resistance of Salmonella serovars in conventional and organic chickens from Louisiana retail stores. J. Food Prot. 2009;72:1165–1172. doi: 10.4315/0362-028x-72.6.1165. [DOI] [PubMed] [Google Scholar]
- Li K., Lemonakis L., Glover B., Mortiz J., Shen C. Impact of built-up-litter and commercial antimicrobials on Salmonella and Campylobacter contamination of broiler carcasses processed at a pilot mobile poultry-processing unit. Front Vet. Sci. 2017;4:1–8. doi: 10.3389/fvets.2017.00088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majowicz S.E., Musto J., Scallan E., Angulo F.J., Kirk M., O’Brien S.J., Jones T.F., Fazil A., Hoekstra R.M. The global burden of Nontyphoidal Salmonella gastroenteritis. Clin. Infect. Dis. 2010;50:882–889. doi: 10.1086/650733. [DOI] [PubMed] [Google Scholar]
- McKee S. American Meat Science Association; Kearney, MO: 2012. Salmonella control in poultry processing. American meat ass. 65th Annual Reciprocal meat Conference; pp. 1–4. [Google Scholar]
- Mezal E.H., Sabol A., Khan M.A., Ali N., Stefanova R. Isolation and molecular characterization of Salmonella enterica serovar Enteritidis from poultry house and clinical samples during 2010. Food Microbiol. 2014;38:67–74. doi: 10.1016/j.fm.2013.08.003. [DOI] [PubMed] [Google Scholar]
- Moore A., Nannapaneni R., Kiess A., Sharma C.S. Evaluation of USDA approved antimicrobials on the reduction of Salmonella and Campylobacter in ground chicken frames and their effect on meat quality. Poult. Sci. 2017;96:2385–2392. doi: 10.3382/ps/pew497. [DOI] [PubMed] [Google Scholar]
- Naravaneni R., Jamil K. Rapid detection of food-borne pathogens by using molecular techniques. J. Med. Microbiol. 2005;54:51–54. doi: 10.1099/jmm.0.45687-0. [DOI] [PubMed] [Google Scholar]
- National Chicken Council (NCC). 2019. Per capita consumption of poultry and livestock, 1965 to estimated 2019, in pounds. Accessed May 2019. https://www.nationalchickencouncil.org/about-the-industry/statistics/per-capita-consumption-of-poultry-and-livestock-1965-to-estimated-2012-in-pounds/ [Google Scholar]
- National Chicken Council (NCC). 2020. Food Safety and Inspection in the U.S. Broiler Chicken Industry. Accessed May 2020. https://www.nationalchickencouncil.org/industry-issues/food-safety/ [Google Scholar]
- Nidaullah H., Abirami N., Shamila-Syuhada A.K., Chua L., Nurul H., Tan T.P., Zainal Abidin F.W., Rusul G. Prevalence of Salmonella in poultry processing environments in wet markets in Penang and Perlis, Malaysia. Vet. World. 2017;10:286–292. doi: 10.14202/vetworld.2017.286-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obe T., Nannapaneni R., Sharma C.S., Kiess A. Rugose morphotype of Salmonella enterica serovar Typhimurium ATCC 14028 exhibits chlorine resistance and strong biofilm forming ability. Int. J. Poult. Sci. 2018;17:295–305. doi: 10.3382/ps/pex346. [DOI] [PubMed] [Google Scholar]
- Oliveira S.D., Santos L.R., Schuch D.M.T., Silva A.B., Salle C.T.P., Canal C.W. Detection and identification of salmonellas from poultry-related samples by PCR. Vet. Microbiol. 2002;87:25–35. doi: 10.1016/s0378-1135(02)00028-7. [DOI] [PubMed] [Google Scholar]
- Olsen J.E., Brown D.J., Madsen M., Bisgaard M. Cross-contamination with Salmonella on broiler slaughterhouse line demonstration by use of epidemiological markers. J. Appl. Microbiol. 2003;94:826–835. doi: 10.1046/j.1365-2672.2003.01911.x. [DOI] [PubMed] [Google Scholar]
- Parveen S., Taabodi M., Schwarz J.G., Oscar T.P., Harter-Dennis J., White D.G. Prevalence and antimicrobial resistance of Salmonella recovered from processed poultry. J. Food Prot. 2007;70:2466–2472. doi: 10.4315/0362-028x-70.11.2466. [DOI] [PubMed] [Google Scholar]
- Perez-Arnedo I., Gonzalez-Fandos E. Prevalence of Campylobacter spp. in poultry in three Spanish farms, a slaughterhouse, and a further processing plant. Foods. 2019;8:111. doi: 10.3390/foods8030111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyrat M.B., Soumet C., marus P., Sanders P. Recovery of Campylobacter jejuni from surfaces of poultry slaughterhouses after cleaning and disinfection procedures: analysis of a potential source of carcass contamination. Int. J. Food Microbiol. 2008;124:188–194. doi: 10.1016/j.ijfoodmicro.2008.03.030. [DOI] [PubMed] [Google Scholar]
- Potter B.D., Marcy J.A., Owens C.M., Slavik M.F., Goodwin H.L., Apple J.K. Impact of performance-based sanitation systems on microbiological characteristics of poultry processing equipment and carcasses as compared with traditional sanitation systems. J. Appl. Poult. Res. 2012;21:669–678. [Google Scholar]
- Rahn K., De Grandis S.A., Clarke R.C., Gala´n J.E., Ginocchio C., Curtiss R., III, Gyles C.L. Amplification of an invA sequence of Salmonella Typhimurium by polymerase chain reaction as a specific method of detection of Salmonella. Mol. Cell. Probes. 1992;6:271–279. doi: 10.1016/0890-8508(92)90002-f. [DOI] [PubMed] [Google Scholar]
- Rasschaert G., Houf K., Godard C., Wildermauwe C., Pastuszak-Frąk M., De Zutter L. Contamination of carcasses with Salmonella during poultry slaughter. J. Food Prot. 2008;71(1):146–152. doi: 10.4315/0362-028x-71.1.146. [DOI] [PubMed] [Google Scholar]
- Rauch H.E., Vosik D., Kariyawasam S., M’ikanatha N., Shariat N.W. Prevalence of group I Salmonella Kentucky in domestic food animals from Pennsylvania and overlap with human clinical CRISPR sequence types. Zoonoses Public Health. 2018;65:7. doi: 10.1111/zph.12506. [DOI] [PubMed] [Google Scholar]
- Reiter M.G.R., Fiorese M.L., Moretto G., López M.C., Jordano R. Prevalence of Salmonella in a poultry slaughterhouse. J. Food Prot. 2007;70:1723–1725. doi: 10.4315/0362-028x-70.7.1723. [DOI] [PubMed] [Google Scholar]
- Russell S.M., Walker J.M. The effect of evisceration on visible contamination and the microbiological profile of fresh broiler chicken carcasses using the Nu-Tech evisceration system or the conventional Streamlined Inspection System. Poult. Sci. 1997;76:780–784. doi: 10.1093/ps/76.5.780. [DOI] [PubMed] [Google Scholar]
- SAS Institute . SAS Institute Inc.; Cary, NC: 2013. SAS User’s Guide: Statistics. Version 9.4. [Google Scholar]
- Schlisselberg D.B., Yaron S. The effect of stainless-steel finish on Salmonella Typhimurium attachment, biofilm formation and sensitivity to chlorine. Food Microbiol. 2013;35:65–72. doi: 10.1016/j.fm.2013.02.005. [DOI] [PubMed] [Google Scholar]
- Shen C., luo Y., Nou X., Bauchan G., Zhou B., Wang Q., Millner P. Enhanced inactivation of salmonella and pseudomonas biofilms on stainless steel by use of t-128, a fresh-produce washing aid, in chlorinated wash solutions. Appl. Environ. Microbiol. 2012;78:6789–6798. doi: 10.1128/AEM.01094-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons M., Fletcher D.L., Carson J.A., Berrang M.E. Recovery of Salmonella from retail broilers by a whole-carcass enrichment procedure. J. Food Prot. 2003;66:446–450. doi: 10.4315/0362-028x-66.3.446. [DOI] [PubMed] [Google Scholar]
- Skyberg J.A., Logue C.M., Nolan L. Virulence genotyping of Salmonella spp. with multiplex PCR. Avian Dis. 2006;50:77–81. doi: 10.1637/7417.1. [DOI] [PubMed] [Google Scholar]
- Smith J., Corkran S., McKee S.R., Bilgili S.F., Singh M. Evaluation of post-chill applications of antimicrobials against Campylobacter jejuni on poultry carcasses. J. Appl. Poult. Res. 2015;24:451–456. [Google Scholar]
- Steel R.G.D., Torrie J.H. McGraw-Hill, Inc; New York, NY: 1980. Principles and Procedures of Statistics: A Biometrical Approach. [Google Scholar]
- Suez J., Porwollik S., Dagan A., Marzel A., Schorr Y.I., Desai P.T., Agmon V., McClelland M., Rahav G., Gal-Mor O. Virulence gene profiling and pathogenicity characterization of non-typhoidal Salmonella accounted for invasive disease in humans. PLoS One. 2013;8:e58449. doi: 10.1371/journal.pone.0058449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tack D.M.E., Marder P., Griffin P.M., Cieslak P.R., Dunn J., Hurd S., Scallan E., Lathrop S., Muse A., Ryan P., Smith K., Tobin-D’Angelo M., Vugia D.J., Holt K.G., Wolpert B.J., Tauxe R., Geissler A.L. Preliminary incidence and trends of infections with pathogens transmitted commonly through food — foodborne diseases active surveillance network, 10 U.S. sites, 2015–2018. MMWR Morb. Mortal Wkly. Rep. 2019;68:369–373. doi: 10.15585/mmwr.mm6816a2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarabees R., Elsayed M.S.A., Shawish R., Basiouni S., Shehata A.A. Isolation and characterization of Salmonella Enteritidis and Salmonella Typhimurium from chicken meat in Egypt. J. Infect. Dev. Ctries. 2017;11:314–319. doi: 10.3855/jidc.8043. [DOI] [PubMed] [Google Scholar]
- Trampel D.W. Recovery of Salmonella from water, equipment, and carcasses in Turkey processing plants. J. Appl. Poult. Res. 2000;9:29–34. [Google Scholar]
- United States Department of Agriculture, Food Safety Inspection Service (USDA-FSIS). 2016. Serotypes Profile of Salmonella Isolates from Meat and Poultry Products, January 1998 through December 2014. Accessed May 2019. https://www.fsis.usda.gov/wps/wcm/connect/3866026a-582d-4f0e-a8ce-851b39c7390f/Salmonella-Serotype-Annual-2014.pdf?MOD=AJPERES. [Google Scholar]
- United States Department of Agriculture, Food Safety Inspection Service (USDA-FSIS). 2017. Table of safe and suitable ingredients: antimicrobial Update 5/25/17. FSIS directive 7120.1 Revision 36. Accessed Aug. 2017. https://www.fsis.usda.gov/wps/wcm/connect/24346cbd-ad28-4223-8db1-55f067ce3879/7120.1-Antimicrobials.pdf?MOD=AJPERES. [Google Scholar]
- United States Department of Agriculture, Food Safety Inspection Service (USDA-FSIS). 2017. Isolation and identification of Salmonella from meat, poultry, pasteurized egg, and siluriformes (fish) products and carcass and environmental sponges. Effective date: 01/02/17. Revised. Accessed Jan. 2019. https://www.fsis.usda.gov/wps/wcm/connect/700c05fe-06a2-492a-a6e1-3357f7701f52/MLG-4.pdf?MOD=AJPERES. [Google Scholar]
- Wideman N., Bailey M., Bilgili S.F., Thippareddi H., Wang L., Bratcher C., Sanchez-Plata M., Singh M. Evaluating best practices for Campylobacter and Salmonella reduction in poultry processing plants. Poult. Sci. 2016;95:306–315. doi: 10.3382/ps/pev328. [DOI] [PubMed] [Google Scholar]
- Yang X., Huang J., Wu Q., Zhang J., Liu S., Guo W., Cai S., Yu S. Prevalence, antimicrobial resistance and genetic diversity of Salmonella isolated from retail ready-to-eat foods in China. Food Control. 2016;16:50–56. [Google Scholar]
- Zhang L., Ren T., Qiao M., Huang T., Xia X. The reduction of Salmonella on chicken skin by the combination of sodium dodecyl sulfate with antimicrobial chemicals and coating wax microemulsions. Poult. Sci. 2019;98:2615–2621. doi: 10.3382/ps/pez008. [DOI] [PubMed] [Google Scholar]