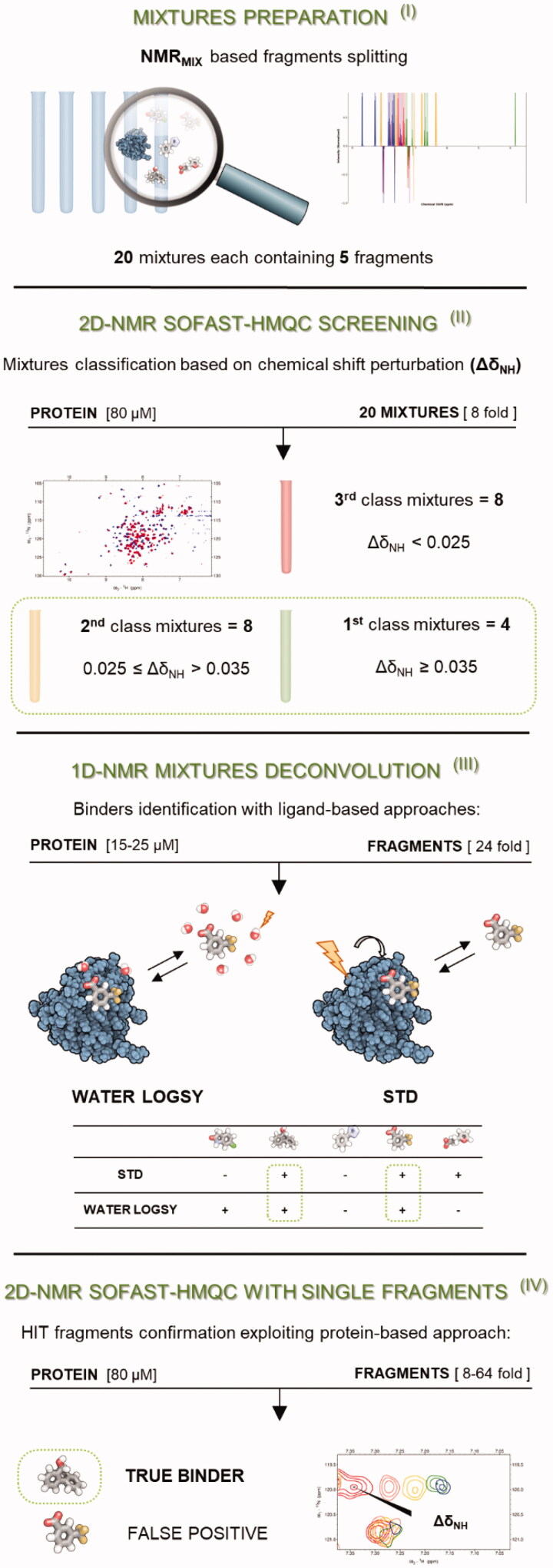
Figure 3.
Schematic representation of the cross-validated NMR protocol followed for the initial screening of our small library containing 100 fragments. First (I), the fragments were divided into 20 mixtures each containing 5 molecules, exploiting NMRmix software37. Then (II), the protein-based experiment SOFAST-HMQC39 was recorded both in the presence and in the absence of the different mixtures, which were classified into three groups depending on their ΔδNH values. Then (III), ligand-based experiments were recorded on the twelve best mixtures to identify the potential binders in every mixture. Finally (IV), the binding of selected molecules was validated exploiting SOFAST-HMQC experiments, collected individually for each fragment.