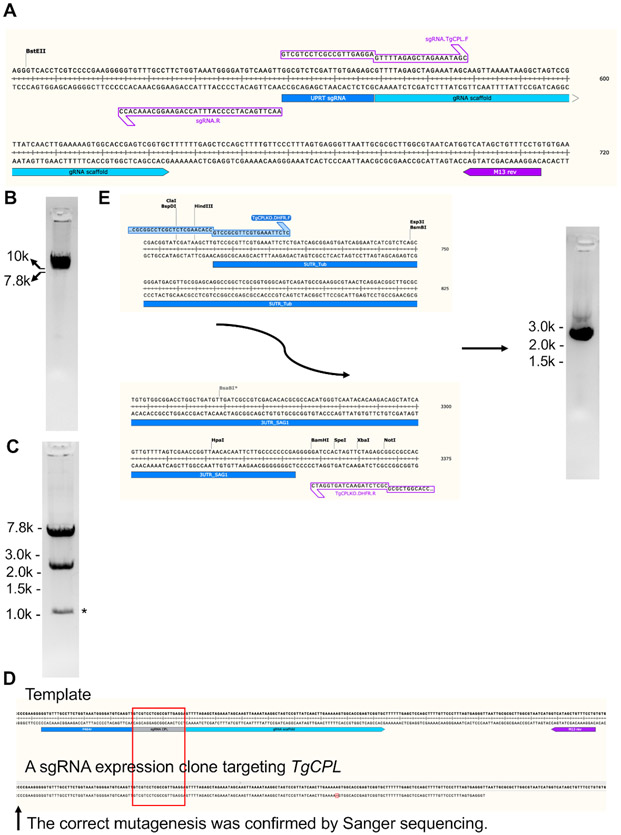
Figure 3: Generation of the plasmid construct expressing sgRNA targeting TgCPL and production of a repair template for TgCPL deletion.
(A) The original pSAG1-Cas9-sgRNA-UPRT plasmid23 was modified via a site-directed mutagenesis kit for replacement of the sgRNA targeting the TgUPRT gene to TgCPL. The sgRNA coding region is enlarged to show areas to which the primers anneal. After PCR, the mutated plasmid was linearized and loaded into a 1% agarose gel for verification of successful amplification, followed by gel extraction. (B) The gel image of the PCR-amplified linearized sgRNA expression construct. (C) After gel-extraction, the PCR product was circularized and subsequently transformed into E. coli. The clones containing the expected plasmids were screened by restriction endonuclease digestion and DNA sequencing. The band sizes after DNA digestion were 7.2 bp and 2.4 kb. The band generated by nonspecific cleavage from endonucleases is labeled by asterisk. (D) The M13 reverse primer labeled in the figure was used to sequence the mutated guide RNA region within the generated TgCPL-targeting sgRNA expression vector. The sequenced DNA region was aligned to the plasmid template to confirm successful mutagenesis. (E) In this study, 50 bp homologous regions matching to the 5'- and 3'-UTRs of TgCPL were engineered into the primers for amplification of the repair template and flanked at the 5'- and 3'-ends of the pyrimethamine resistance cassette by PCR, respectively. Agarose gel electrophoresis was used to verify the correct size of the PCR product before gel extraction. The expected size of the repair template is ~2.7 kb. Usually, 5-6 μg of repair template can be obtained from 200 μL of PCR reaction.