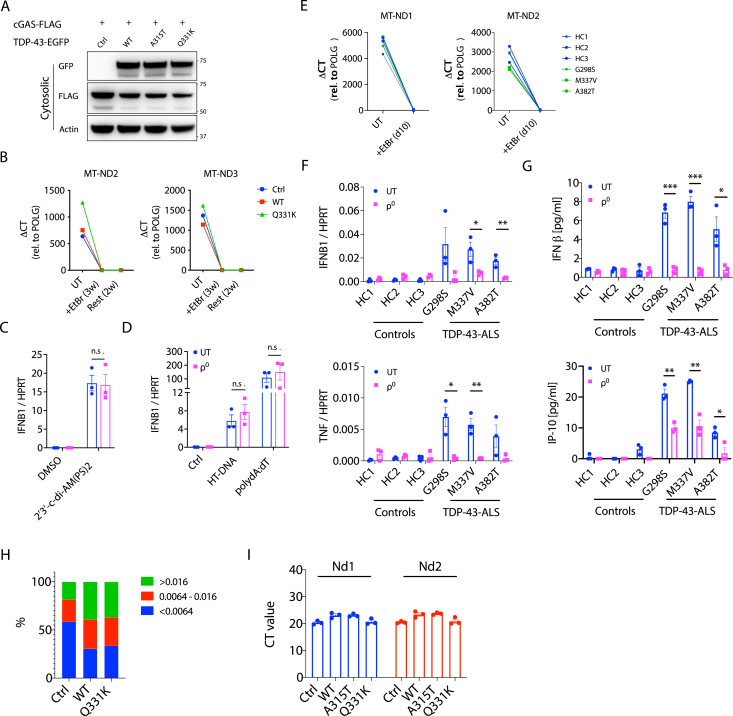
Figure S2.
mtDNA Is a DAMP that Activates cGAS/STING Signaling, Related to Figure 2
(A) Representative western blot of FLAG-cGAS immunoprecipitation from Figure 2A. (B) Representative qPCR analysis of mitochondrial DNA (mtDNA) depletion from THP-1 cells over three weeks treatment in ethidium bromide (EtBr) and then two weeks after removing EtBr. To indicate the depletion, mitochondrial genes (MT-ND2 and MT-ND3) were normalized to that of nuclear gene POLG as ΔCT and compared to untreated cells (UT). (C) IFNB1 expression, measured by qPCR, in UT and ρ0 THP-1 cells in response to stimulation with 2′3′-c-di-AM(PS)2(Rp, Rp) (20 μM) or DMSO as solvent control for 4hrs or (D) poly(dA:dT) (1 μg/ml), HT-DNA (2 μg/ml) and equivalent Lipofectamine as control (Ctrl) for 6hrs. (E) mitochondrial genes (MT-ND1 and MT-ND2) were depleted in iPSC-MNs treated with EtBr for 10 days. (F) Gene expression of IFNB1 and TNF, and (G) IFNβ and IP-10 production were diminished in ρ0 iPSC-MNs from TDP-43-ALS patients. (H) Quantification of mtDNA nucleoid size for imaging data in Figure 2D. Nucleoid sizes were divided into three groups; < 0.0064 mm3, 0.0064-0.016 mm3 and > 0.016 mm3. (I) No difference in copies of mitochondrial DNA (Nd1 and Nd2) was observed in total cell lysates of MEFs transfected with WT or mutant TDP-43 (A315T and Q331K). Data are mean ± SEM from 3 independent experiments. P values were calculated using two-way ANOVA. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, n.s. not significant.