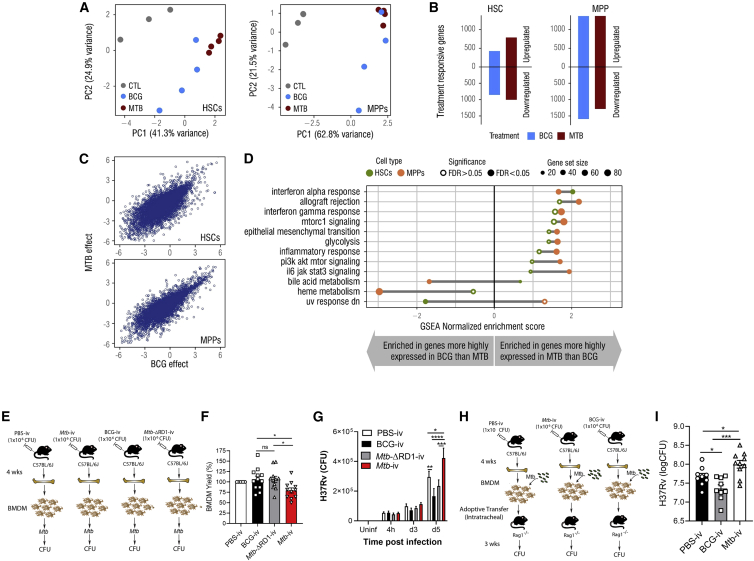
Figure 2.
Mtb-i.v. Imprints a Unique Transcriptional Signature in HSCs and Detrimentally Trains BMDMs
(A) Principal-component analysis (PCA) of gene expression of HSCs and MPPs from the BM of PBS, BCG-i.v., and Mtb-i.v. mice after 28 days.
(B) Number of genes differentially up- or downregulated in HSCs and MPPs (FDR < 0.01).
(C) Scatterplots of genome-wide effect sizes in HSCs (top) and MPPs (bottom).
(D) GSEA of DEGs (FDR < 0.1).
(E) BMDM model.
(F) Relative BMDM yield.
(G) CFUs from in vitro Mtb-infected BMDMs (n = 7–10 mice/group).
(H) Adoptive transfer model.
(I) Lung CFUs post-transfer.
In (F) and (I), one-way ANOVA followed by Tukey’s multiple comparisons test was used. In (G), two-way ANOVA followed by Sidak’s multiple comparisons test was used. In (E)–(I), data are pooled from 2–3 independent experiment. See also Figure S2.