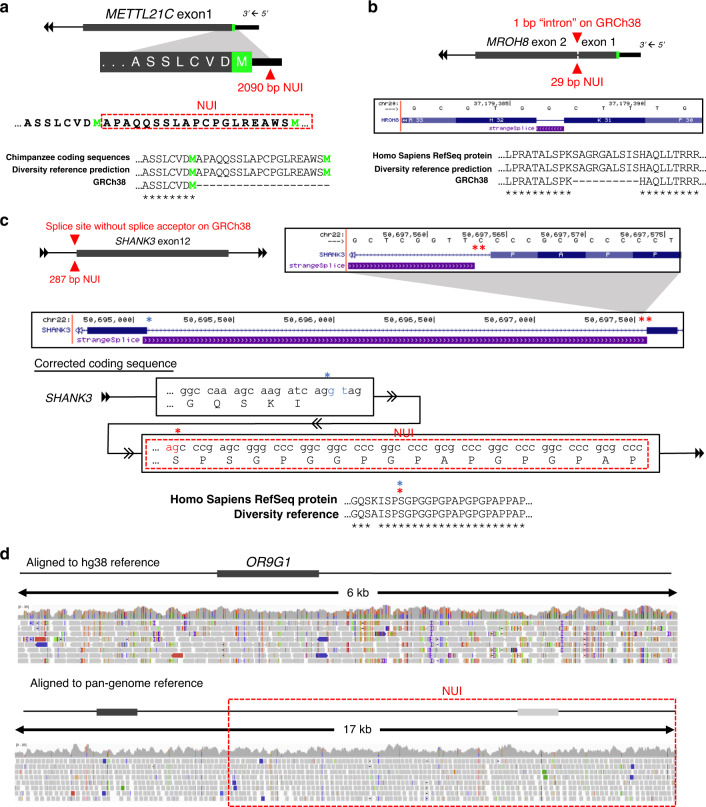
Fig. 3. Examples of NUIs affecting exonic annotations.
a A 2090 bp insertion was identified 4 bp upstream of the annotated start codon based on GRCh38 annotation. Upon integrating the NUI, the open reading frame is extended by 20 amino acids with a new methionine that could now be served as the start codon. Identical coding sequences were identified in the chimpanzee RefSeq protein database. Neon green denotes the annotated and the new start codon methionine. b A 29 bp NUI was identified precisely at that 1 bp “intron” between exon 1 and exon 2 of MROH8. It restores a single reading frame by merging the two exons. The new coding sequence has a match in the human RefSeq protein database. c A 287 bp NUI was located right at the 3′ end (splice acceptor) of the intron between exon 11 and exon 12 of SHANK3. The UCSC genome browser screenshot showed a non-canonical splice site that did not match with the corresponding splice donor. Upon integrating the sequence boxed in red, the bona fide splice acceptor was restored. The full-length coding sequence had an almost identical match to a record in the human RefSeq protein database. Blue asterisk, splice donor; single red asterisk, restored splice acceptor; double red asterisks, original splice acceptor. d An example demonstrating the excessive number of variants caused by missing sequences. This missing sequence also contains an ortholog of OR9G1 that is not present in the GRCh38 core reference. Gene structures are not drawn to scale.