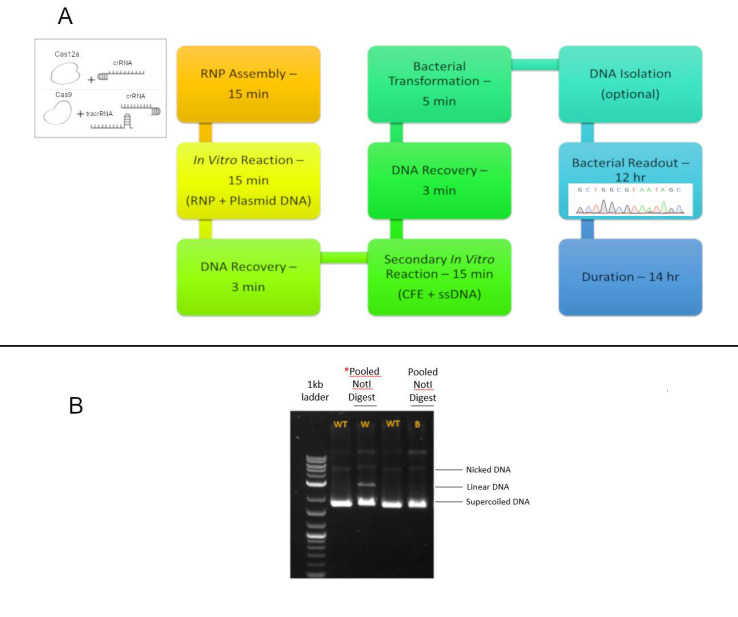
Figure 2.
In vitro gene editing workflow and NOTI digestion assay. (A) In vitro workflow. The workflow starts with the complexation of CRISPR/Cas12 and/or CRISPR/Cas9 with the appropriate crRNA/tracrRNA+crRNA’s to form a ribonucleoprotein (RNP). After formation, the RNP is added to a reaction containing plasmid DNA, incubated for 15 min, and then cleaved DNA is recovered. The DNA is added to a second reaction with cell-free extract, ssDNA oligonucleotide, and undergoes DNA recovery once more. The recovered DNA is transformed into Escherichia coli bacteria, incubated overnight, and bacterial readout is observed after 12 h. (B) Pooled NOTI digestion assay. A NOTI digestion agarose gel is shown with five columns; the first column illustrates the 1 kb ladder and the remaining four columns show pooled white and blue colony samples along with a control, uncut LacZ plasmid. A linear band can be seen in the pooled white colony samples indicating several colonies with an intact NOTI site. The red asterisk signifies that only the white colony pool demonstrated NOTI cleavage. The pooled blue sample shows only wildtype (WT) uncut, plasmid DNA.