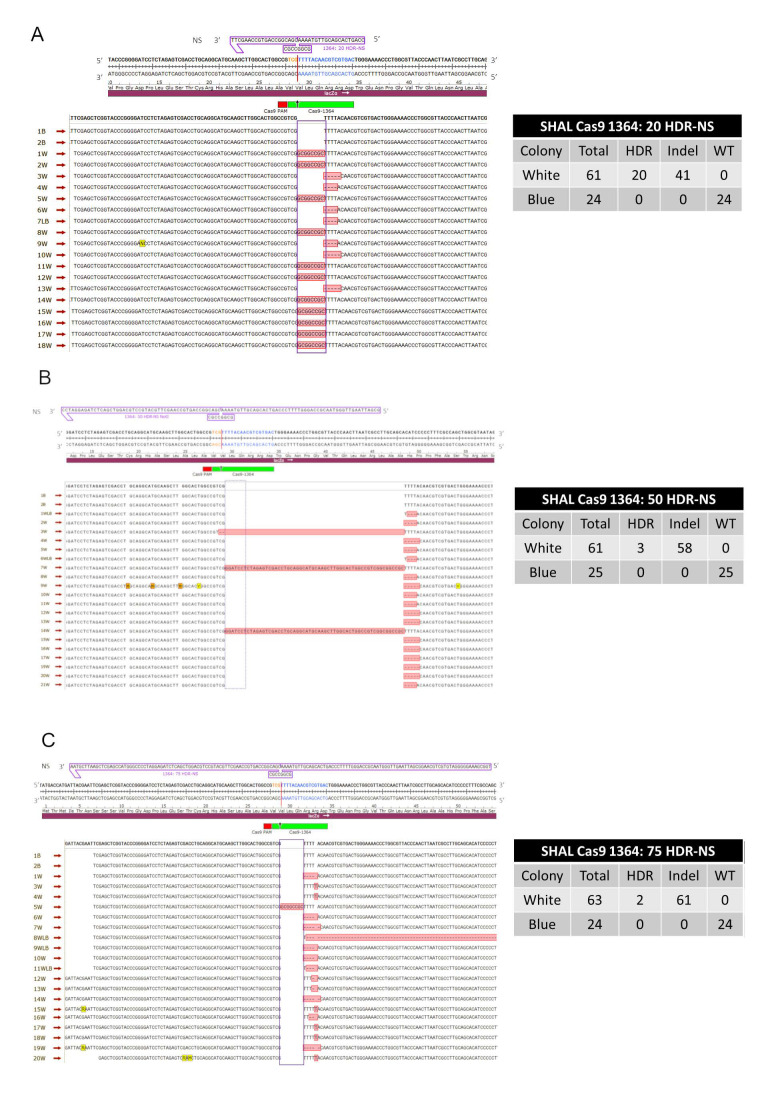
Figure 4.
(A–C) Symmetrical homology arm lengths using CRISPR/Cas9. Each image illustrates the location of Cas9 in green and the PAM site in red. A solid red line indicates the blunt ended cleavage site. (A–C) symmetrical homology arm lengths of 20, 50, and 75 base pairs (48, 108, and 158 bases total). The nonsense symmetrical oligonucleotide with lengths of 20, 50, or 75 base pairs on either side of the cut site is located above the lacZ gene, contains an eight base NOTI site in the center, and is positioned in a 3′–5′ orientation. The purple box highlights the correct integration of a NOTI site and bases highlighted in red outside of the purple box indicate unintended integration or deletion events for each image. The table to the right of the images illustrates the total number of blue and white colonies sequenced and if they were found to harbor homology-directed repair, insertions or deletions (Indels), or a wild-type event. All blue colonies were found to harbor wild-type sequences represented by the two blue colonies (labeled 1B and 2B) in all images. All experiments were conducted in triplicate and the combined data is shown only in the tables present. The remaining sequencing data is shown in Figure S2 and the individual colony numbers for each trial are shown in Table S2.