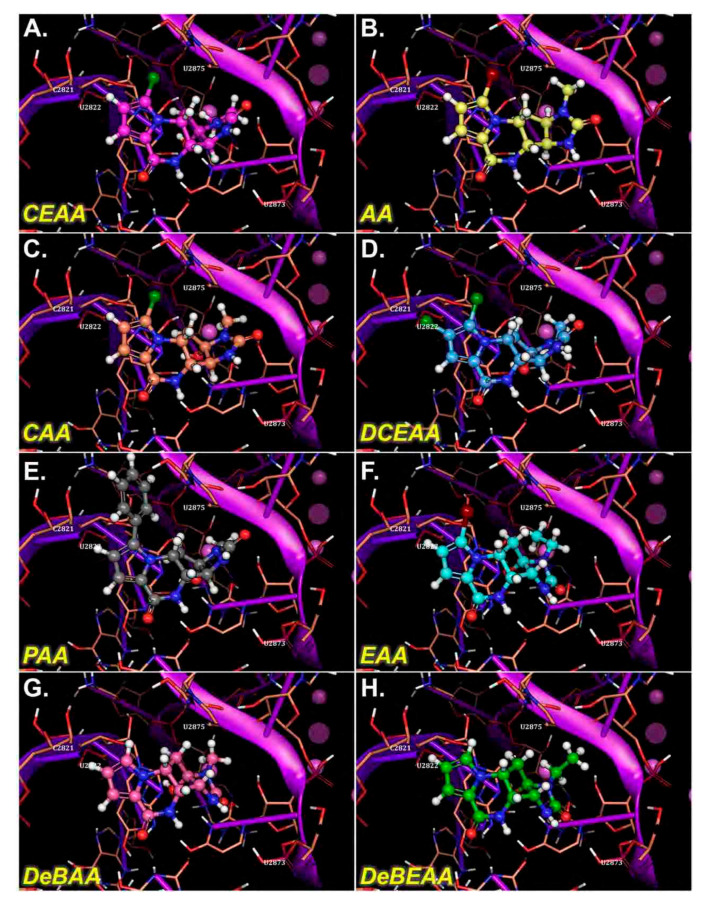
Figure 4.
Three-dimensional docked images for AA, CAA, CEAA, EAA, PAA, DCEAA, DeBAA, and DeBEAA with ribosome’s PTC region (prior to Molecular Mechanics-Generalized-Born Solvent Accessible (MM-GBSA) energy calculations). Docking assigns the compounds pose or position in the PTC. The potent derivatives, CEAA, AA, DCEAA and CAA have a similar orientation in their binding and other less potent derivatives lack matching overlap. As the docking is machine driven to the optimal score possible and alternative positions drop with increasingly positive scores, these orientations illustrate the effect subtle changes have on compound efficacy and orientation. Here, the relevant interactions between ribosome and the AA that form important binding sites are shown. (A) AA is shown bound in the ribosomal PTC, which has good docking pose that matches the X-ray structure (see two-dimensional ligand interaction map for details on binding residues). (B) CEAA shows the critical residues needed for binding to PTC. (C) DCEAA is shown with the PTC region surrounding. (D) CAA has some similar binding characteristics to that of AA on the PTC. (E) PAA is given as bound in the ribosome. (F) EAA has weak binding in the PTC. (G) DeBAA is shown docked into PTC. (H) DeBEAA has weak binding at the PTC.