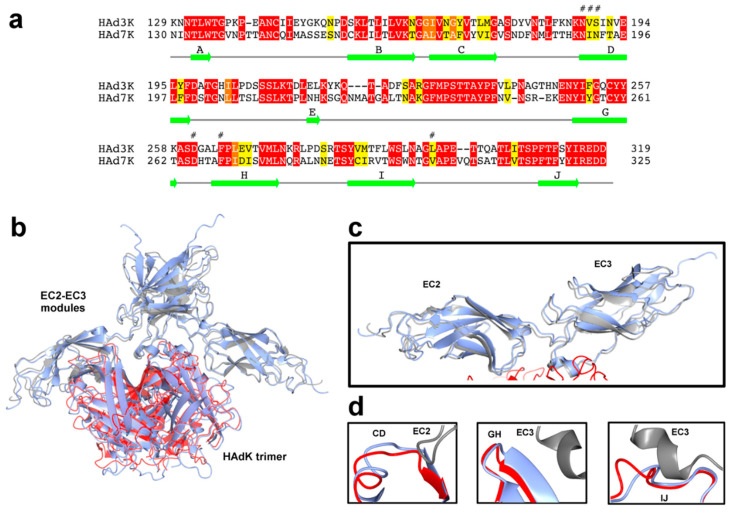
Figure 5.
(a) Pairwise Sequence Alignment is used to identify regions of similarity between HAdV7 and HAdV3 fiber. For each pair of sequences the best global alignment is found using BLOSUM62 as the scoring matrix. A conservation index between 0 and 11 is calculated for each amino acid position on the alignment. Amino acids with a score above 9 are colored from red→orange→yellow corresponding to sequence conservation from high to low. (b) Superposition of HAd7K-(EC23)2 and HAd3K-(EC23)2: EC2-EC3 modules interacting with HAd7K (in red) are colored in grey. HAd3K-(EC23)2 complex is colored in blue. (c,d) Close-up views of superposed EC2-EC3 modules and superposed CD, GH, and IJ loops of HAdK, are shown individually.