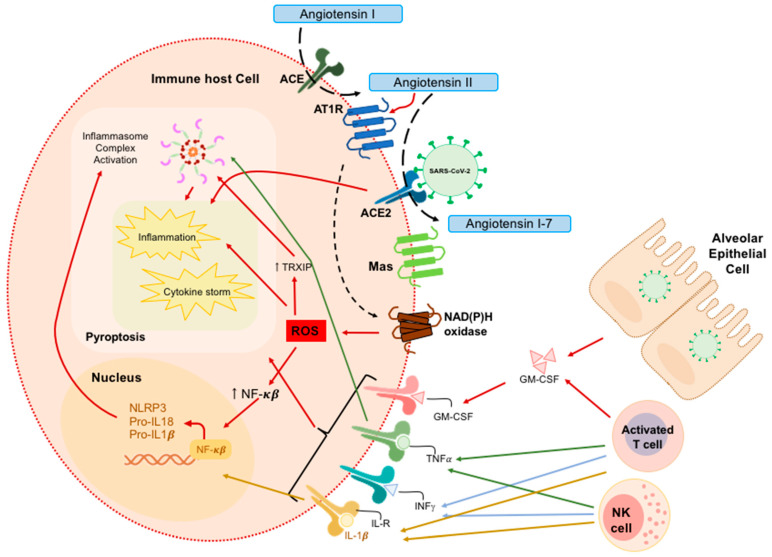
Figure 1.
Molecular pathogenesis of SARS-CoV-2. SARS-CoV-2 virus can bind to specific receptors in host cells, such as alveolar epithelial cells or immune cells, by mediating an inflammatory cascade through inflammasome activation, usually through NLRP3, and by damaging, or even killing, host cells. Immune host cells, like macrophages and monocytes, are activated by the virus on a direct or indirect pathway, and contribute to the host response by causing inflammation and a cytokine storm. Other immune cells, such as NK cells and T-cells, can contribute to the immune response. On the other hand, Ang II is processed by ACE2 into Ang I-7. Ang II contributes to ROS production in an NAD(P)H-dependent mechanism thanks to the ACE2–Ang-(1–7)–Mas axis. AT1R mediates the production of ROS through a mechanism dependent on NADH and NADPH oxidases. ROS contributes to nuclear factor-kB (NF-κB) and TXNIP overexpression. NF-κB increases the expression of NLRP3, pro-IL-18, and pro-IL-1β whilst TXNIP modulates the structure of NLRP3, thereby allowing NLRP3 inflammasome assembly and facilitating pro-caspase-1 (pro-casp-1) autocleavage. Arrows denote activation. ACE2: angiotensin-converting enzyme 2; GM-CSF: Granulocyte-macrophage colony-stimulating factor; TNF: Tumor Necrosis Factor; INFγ: Interferon gamma; IL-R: Interleukin Receptor; AT1R: Angiotensin type 1; NADPH oxidase: nicotinamide adenine dinucleotide phosphate oxidase; ROS: reactive oxygen species. TXNIP: thioredoxin interacting/inhibiting protein. This figure is adapted from the work of Merad et al. [33].