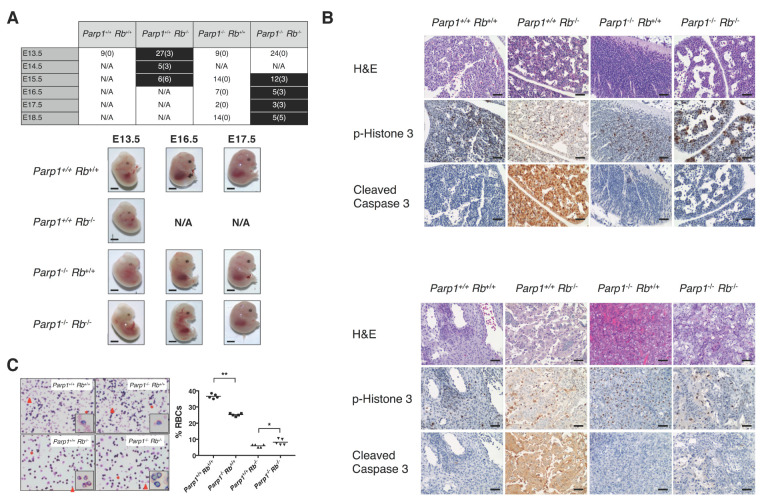
Figure 1.
Genetic deletion of Parp1 restores the phenotypic alterations induced by E2F1 hyperactivation in Rb-/- embryos. (A) Summary of embryos analysed from each genotype at different gestational ages. Non-viable embryos are denoted in brackets. Timepoints affected by embryonic lethality are highlighted in black. While Parp1+/+ Rb−/− mice display embryonic lethality at circa E13.5, Parp1−/− Rb−/−mice show a delay until E16.5 (p < 0.05). Lower panel exhibits the macroscopic differences of the different genotypes at E13.5, E16.5 and E17.5. Rb+/- embryos at E16.5 and E17.5 are not displayed due to embryo resorption. Solid arrowheads highlight functional vascularization and liver integrity at E16.5 and E17.5 in Parp1−/− Rb−/−, resembling those of other viable genotypes Scale = 1 mm. (B) Histological analysis of foetal liver tissue at E13.5. Unlike Rb-deficient mice, double knock-out embryos do not display high levels of cleaved caspase 3, a hallmark of apoptosis. Staining of phospho-histone 3 reveals a significant rate of proliferation, not much different than that of wild-type or Parp1 knock-out mice. Scale (40×) = 25 μm. Histological analysis of placental tissue revealed massive apoptosis in cells lacking retinoblastoma, while Parp1−/− Rb−/− placentas displayed a slight increase in cleaved caspase 3 staining compared to samples from wild-type or Parp1-/- mice. Scale (40×) = 25 μm. (C) Counting of enucleated red blood cells (RBCs) showed a drop-in cell numbers of both retinoblastoma-deficient groups that is not observed Parp1-/- Rb-/- mice (* p < 0.05; ** p < 0.002). Scale (40×) = 25 μm.