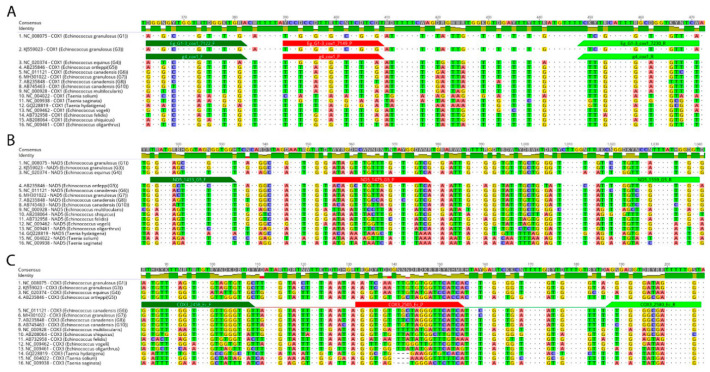
Figure 1.
Location of Echinococcus granulosus sensu lato polymorphic genome regions within COX1, COX3 and NAD5 genes used for primer and probes design. In-silico identification of polymorphic regions in aligned mitochondrial DNA sequences to differentiate the E. granulosus sensu lato species E. granulosus sensu stricto (G1–3): (A), E. equinus (G4); (A), E. ortleppi (G5); (B) and E. canadensis (G6–8, G10); (C) from other Echinococcus spp. and Taenia spp. Differences between aligned sequences are highlighted by different background colours of respective nucleotides. Dots represent nucleotides identical in all aligned sequences. Dark-green bars show the positions of the respective forward primer, red bars annotate the positions of the respective probes, and light-green bars indicate the positions of the reverse primer. The name of each aligned sequence consists of NCBI nucleotide database accession number, followed by gene name and the name of Echinococcus species. G: genotype.