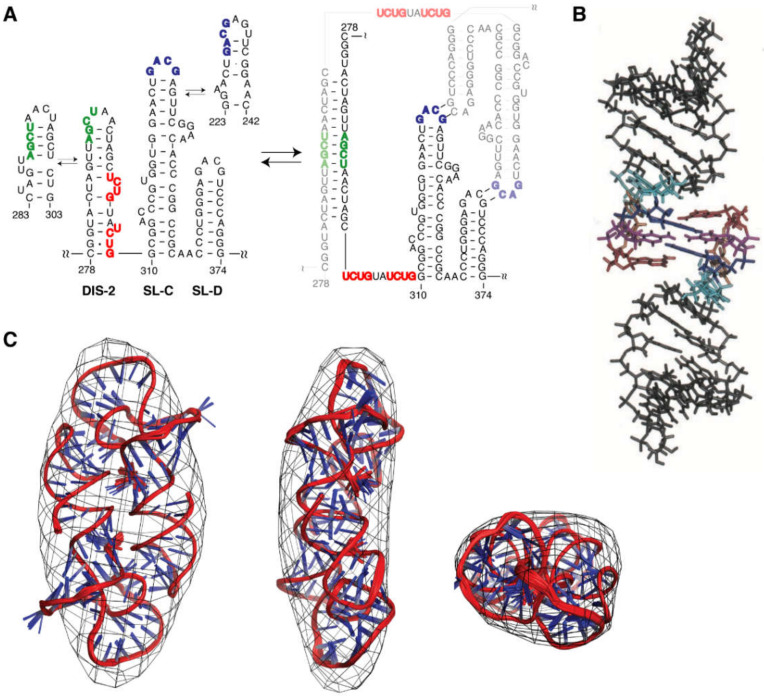
Figure 13.
Dimerization-dependent structure of the MoMuLV minimal packaging signal. (A) Left: Secondary structure of monomeric mΨ and the base pairings observed at equilibrium for DIS-2 and SL-C. Right: Base pairings in the dimeric form of mΨ. The palindromic sequences required for dimer formation are shown in green (DIS-2) and blue (SL-C and SL-D). NC-binding UCUG elements are shown in red. (B) The NMR structure of the 18-nt SL-D kissing duplex. (C) Superposition of the cryo-electron tomography densities and the NMR ensemble structures of [ΨCD]2. Panels (A,C) reproduced from [315], and panel (B) from [316], with permission.