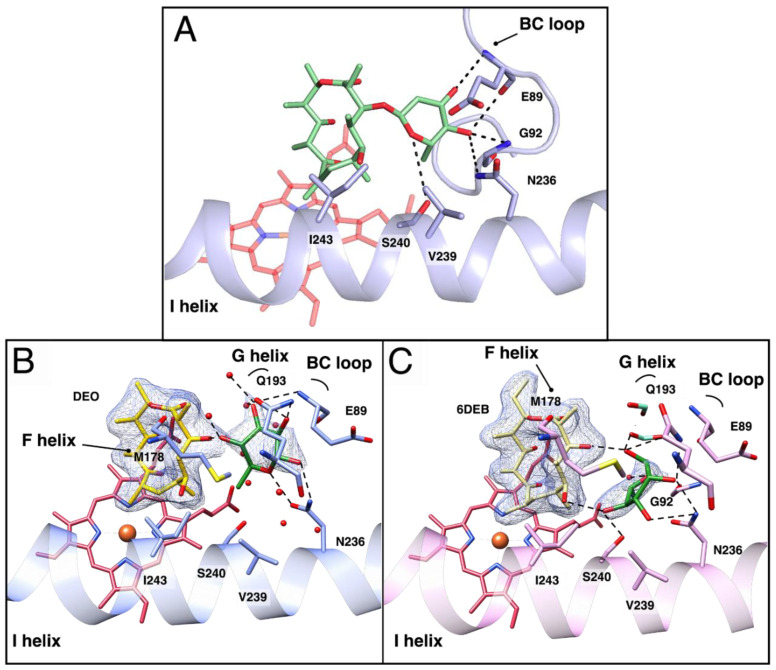
Figure 4.
In silico docking of L-O-DEO and structures of OleP-aglycones bound to L-rhamnose. (A). Zoom on the active site of closed OleP (blue) into which L-O-DEO (lime green sticks) was computationally docked using AutoDock Vina 1.1.2. Residues forming direct hydrogen bonds (dashed lines) with the olivosyl moiety of L-O-DEO, placed at a distance ranging between 2.4 and 4 Å, are labeled. (B,C). Close up views of the active site OleP–DEO-rhamnose (panel B, blue) and OleP–6DEB–rhamnose (panel C, pink). Residues and solvent molecules (water, red spheres; formate ions, aquamarine sticks) within 5 Å from L-rhamnose are displayed. Dashed lines represent hydrogen bonds. Secondary structural elements and amino acids are labeled. In both panels, the electron density map (2Fo–Fc) contoured at 1 σ around L-rhamnose (green sticks) and DEO (yellow sticks, panel B) or 6DEB (khaki sticks, panel C) is shown as a blue mesh.