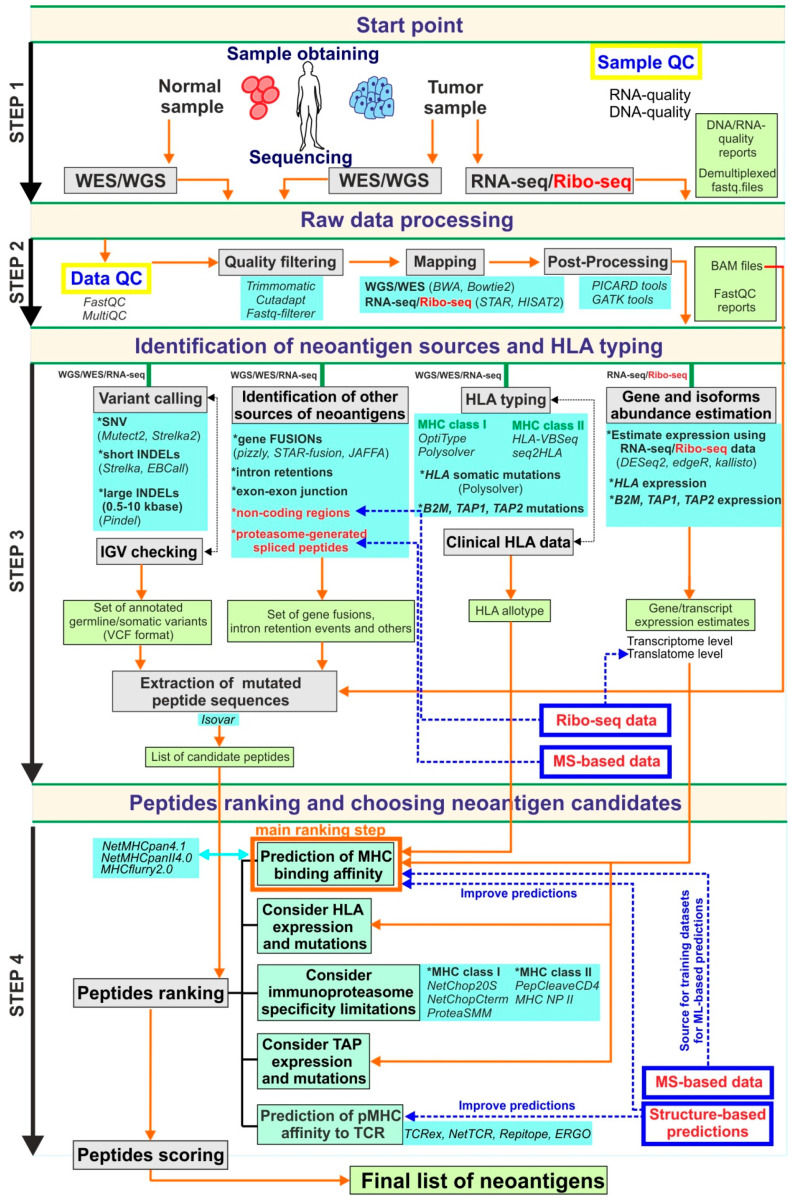
Figure 1.
The schematic description of the possible ideal genomics-centric pipeline for neoantigen identification. In this scheme, the pipeline is formally split into four steps. Step 1 is related to sample obtaining, DNA/RNA-extraction, libraries preparation and high-throughput sequencing. Step 2 is associated with raw NGS data processing, quality-filtering and obtaining aligned to reference reads in an appropriate format suitable for downstream analysis. The aim of Step 3 is to obtain all possible information from processed NGS including all variants set, HLA allotype, expression estimations, as well as, candidate peptide sequences that further to prioritization procedures of Step 4. At the final Step 4 candidate peptide list obtaining based on identified variants used for peptide ranking using mainly MHC binding estimators. Additional options such as TCR-pMHC binding affinity scoring, TAP-transport, etc., should be considered during the prioritization step. Grey boxes are reflected main procedures that should be done in each step; light green boxes contain data that should be generated on each step; light blue boxes present possible tools that could be applied for the corresponding procedure; blue boxes with red text (as well as red text alone) are related to additional approaches that could add value to this pipeline; the orange arrows show the information flow through pipeline workflow; the blue dashed arrows highlight the steps that could be improved by additional approaches.