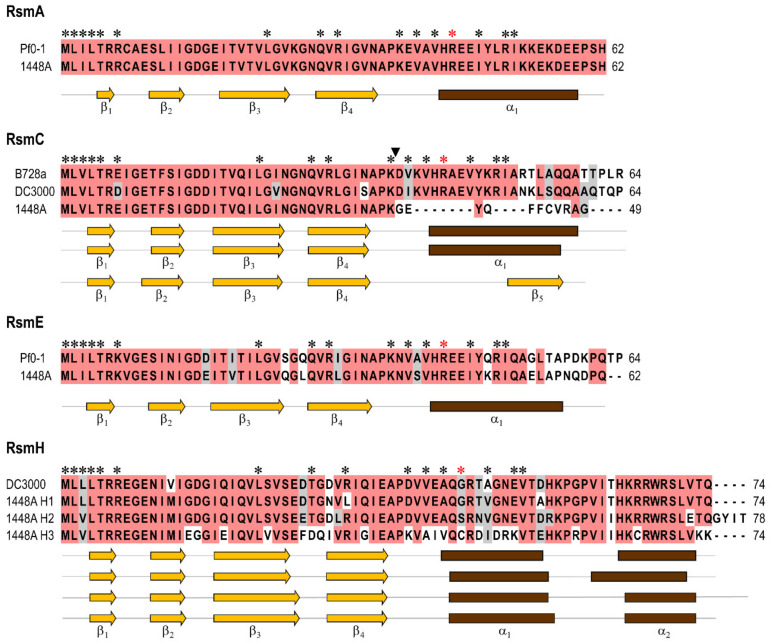
Figure 1.
Sequence and structural conservation of the deduced products of the rsm homologues from P. amygdali pv. phaseolicola 1448A. All proteins were aligned using ClustalW, and identical or similar residues, shared by at least 60% of the sequences in multiple alignments, are shaded in red and grey, respectively. Dashes indicate gaps introduced to maximize the alignment. Asterisks indicate residues that are important for the interaction between RsmA or RsmE with RNA and for the regulation of several phenotypes [67,68,69], with the red asterisk indicating a critical Arg44 residue in RsmA and RsmE; the corresponding residues in RsmC and RsmH were marked from a sequence alignment with RsmA and RsmE. The black arrowhead indicates the point where an insertion of ISPsy17 interrupts the rsmC coding sequence in strain Pph 1448A. Secondary structures were predicted with JPred4 and are shown below the alignments and in the same vertical order than the respective sequences; secondary structures were identical for RsmA and RsmE proteins, and so only one is shown for each. Abbreviations (accession numbers): Pf0-1, P. fluorescens Pf0-1 (RsmA, WP_002554426; RsmE, WP_003179932); 1448A, P. amygdali pv. phaseolicola 1448A; B728a, P. syringae pv. syringae B728a; DC3000, P. syringae pv. tomato DC3000; accession numbers for 1448A, B728a and DC3000 proteins are indicated in Table S3.