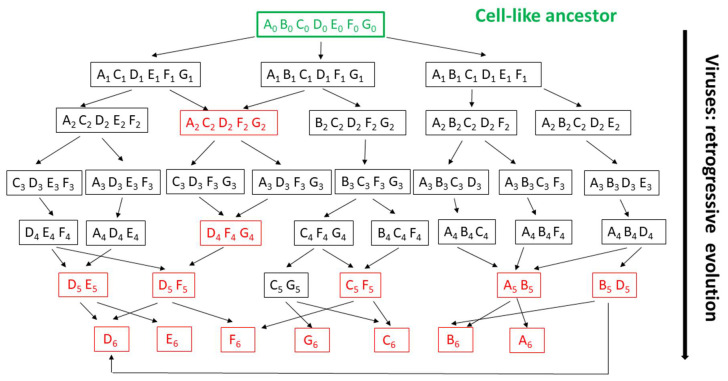
Figure 3.
The “virus late” hypothesis: illustration of the intractable evolutionary scenarios resulting from random gene/function losses. A toy virus world is represented, starting from a hypothetical ancestral cell-like organism (level 0, w/o extant representative). Each box contains the abstract gene content inherited by a given virus (family) from its immediate ancestor. Red “genomes” indicate viruses with shared gene contents albeit possibly resulting from distinct evolutionary pathways. Random gene losses lead to very diverse overlaps of gene assortments (as in viruses) or to situations where no single “core gene” is shared by all virus family (here starting at level 3), as observed in actual viral genomes (in particular small ones). Individual genes recurring in multiple combinations (families) or ultimately remaining in the smallest genomes (level 6) are not more characteristic of the parasitic lifestyle than less ubiquitous ones. In addition, genes shared by more families than other (such as D) may not be better phylogenetic markers than others, as they could have been inherited from different ancestors (DE, DF, BD). Their polyphyly will not be detected if some of their above ancestors are extinct or unknown. This graph illustrates the difficulty of reconstructing the deep phylogeny of viruses beyond the immediate family level both due to the capacity of random gene losses enjoyed by obligate intracellular parasites and the lack of associated free-living organisms to be used as references.