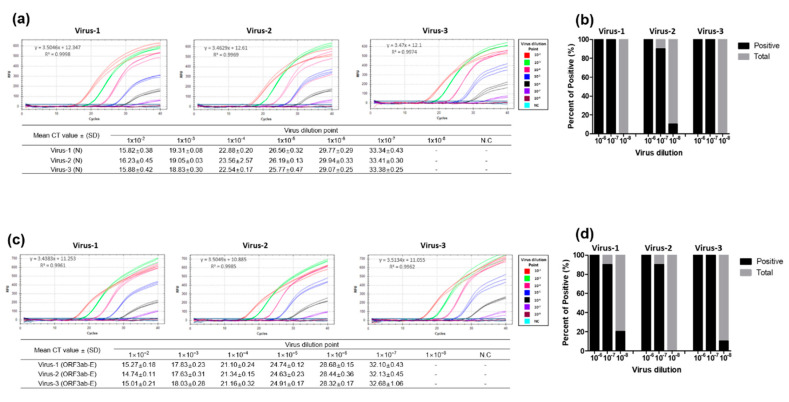
Figure 5.
Sensitivity evaluation of the designed PNA-mediated RT-qPCR assay and the enhanced universal RT-qPCR method for detection using intact SARS-CoV-2 RNA. The evaluation was performed using RNA extracts from cell-propagated SARS-CoV-2 viruses isolated from patients diagnosed with COVID-19. Intact viral RNA extracts were ten-fold serially diluted (10−2 to 10−8) and processed for the detection of SARS-CoV-2 using the designed primers for N (a) and ORF3ab-E genes (b). RT-qPCR-positive amplification was determined from the mean cycle threshold value for each RNA dilution point. (c,d) Limit of detection was assessed using 10−6, 10−7, and 10−8 dilutions in ten repetitions. N.C: negative control; Ct: cycle threshold; SD: standard deviation; “-”: not determined.