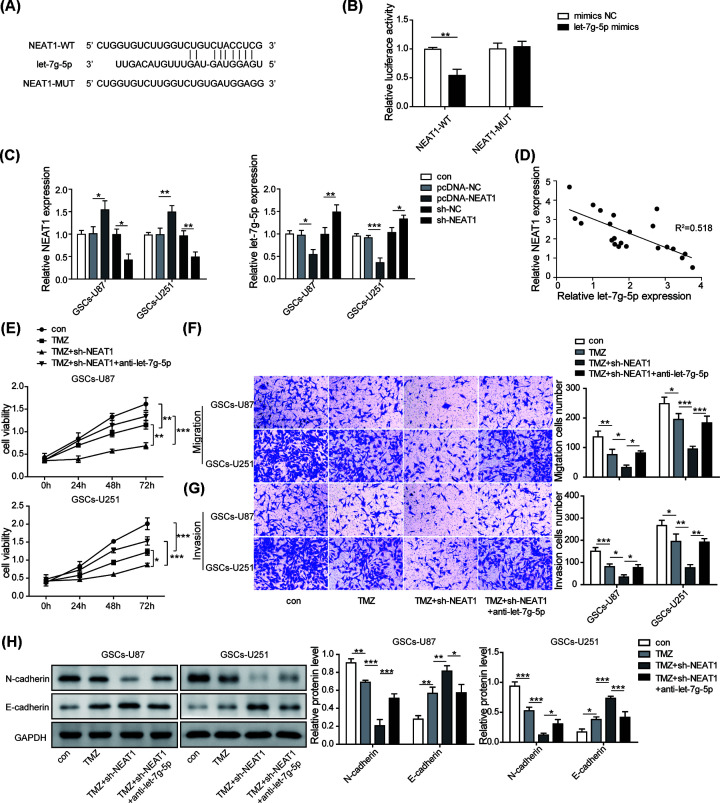
Figure 4. let-7g-5p negatively affected the functions of NEAT1.
(A) Bioinformatics method (Starbase) was used to predict the potential binding site between NEAT1 and let-7g-5p. (B) Dual-luciferase reporter assay was used to evaluate targeted relationship. (C) qRT-PCR was used to test the regulatory correlation between NEAT1 and let-7g-5p on expression level. (D) The expression correlation of NEAT1 and let-7g-5p in clinical samples was analyzed by Pearson analysis. (E) CCK-8 assay was used to explore that the effect of let-7g-5p down-regulation on NEAT1-mediated GSCs resistance to TMZ. (F,G) Transwell assay was performed to assess the roles of let-7g-5p down-regulation on the capacities of migration and invasion of GSCs on the basis of TMZ+shNEAT1 co-treatment. (H) Western blot assay was used to evaluate the roles of let-7g-5p down-regulation on the levels of E-cadherin and N-cadherin mediated by TMZ and shNEAT1 co-treatment. Data were presented as mean ± SD; *P<0.5, **P<0.01, ***P<0.001.