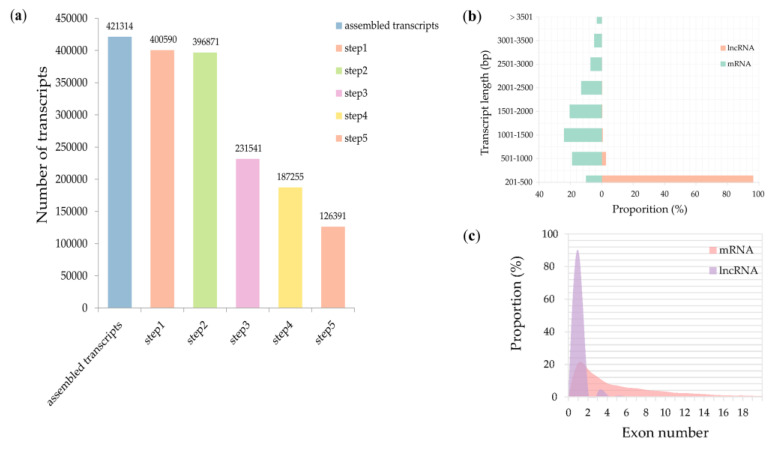
Figure 1.
Identification and characterization of long noncoding RNAs (lncRNAs). (a) Statistics of candidate lncRNAs. Step 1: select transcripts with exon number ≥ 1 and filter out numerous single-exon transcripts by FPKM (fragments per kilobase of exon per million fragments mapped) value < 0.5. Step 2: select transcripts with length > 200 bp. Step 3: filter out transcripts overlapped with the exon region of the database annotation. Step 4: retain the transcripts characterized by FPKM value ≥ 0.5 (for single-exon transcripts, FPKM value ≥ 2). Step 5: remove the transcripts that failed to pass the protein-coding-score test. (b) Comparison of the transcript length between lncRNAs and mRNAs; (c) exon number distribution of lncRNAs and mRNAs.