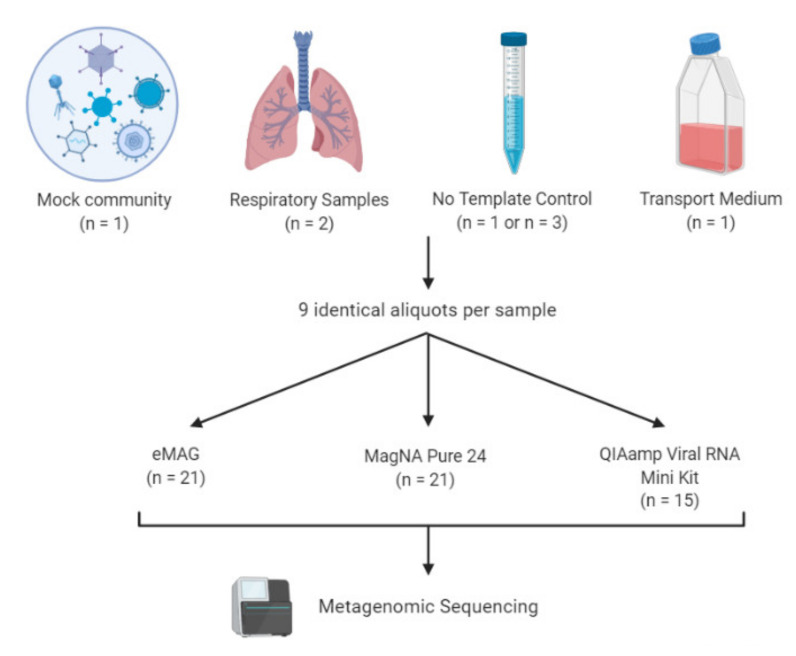
Figure 1.
Overview of the study design. A mock virome containing five viruses isolated from respiratory samples representative of a wide range of virus characteristics (adenovirus 31, respiratory syncytial virus A, herpes simplex virus 1, influenza A virus, and rhinovirus) was prepared. Human clinical samples were obtained from hospitalized patients (one bronchoalveolar lavage positive for herpes simplex virus 1 (HSV-1) and one nasopharyngeal aspiration positive for respiratory syncytial virus A (RSV-A). No template controls (NTCs) and transport medium samples were implemented in the process to control for sample and kitome cross-contamination. For the automatic extractors, NTCs were interspersed between the samples of each batch (n=3 per batch), whereas for the manual method only 1 NTC was included per batch (n = 1). To assess the reliability and reproducibility of the experimental results, the extractions and next generation sequencing (NGS) workflow were set up in triplicate (for the mock and respiratory samples), using the same amount of sample input. Finally, libraries were sequenced in the same run with Illumina NextSeq 500 ™ using a 2 x 150 paired-end (PE) high-output flow cell.