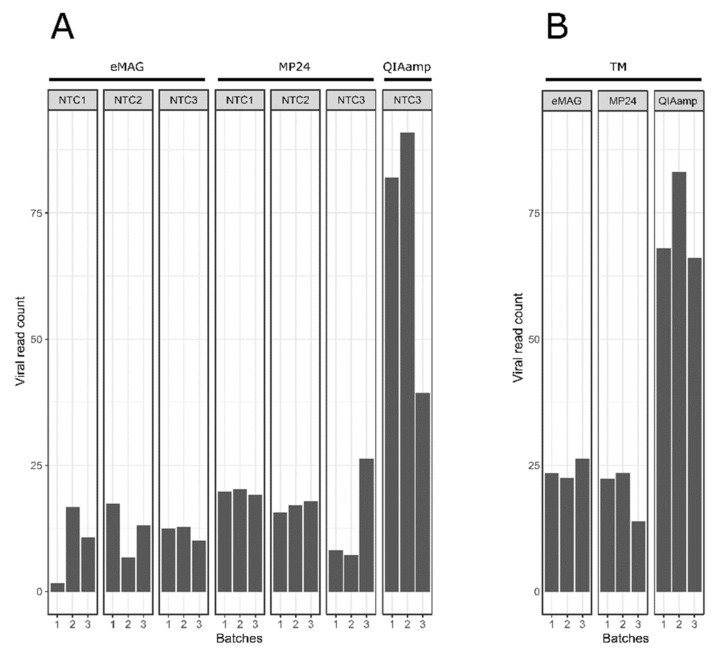
Figure 6.
Proportion of Kitome contained in each triplicate from the different extraction methods (A) in NTC and (B) in the transport medium. Bar plot showing the sum of the viral read count normalized in log10(RPM), associated with reagent contamination (i.e., reads associated with other viruses than the targeted viruses: kitome) for each NTC and TM compared between different extraction methods. During automated extractions, NTCs were interspersed between samples (i.e., 3 NTCs per batch). For manual extractions, only one NTC was added. In addition to the NTC, a transport medium was added. Analyses were performed at the family taxonomy level. A sample was considered to be positive for a particular virus when the log10(RPM) of this virus exceeded 1. HSV: herpes simplex virus; RSV: respiratory syncytial virus; NTC: no template control; TM: transport medium; RPM: reads per million.