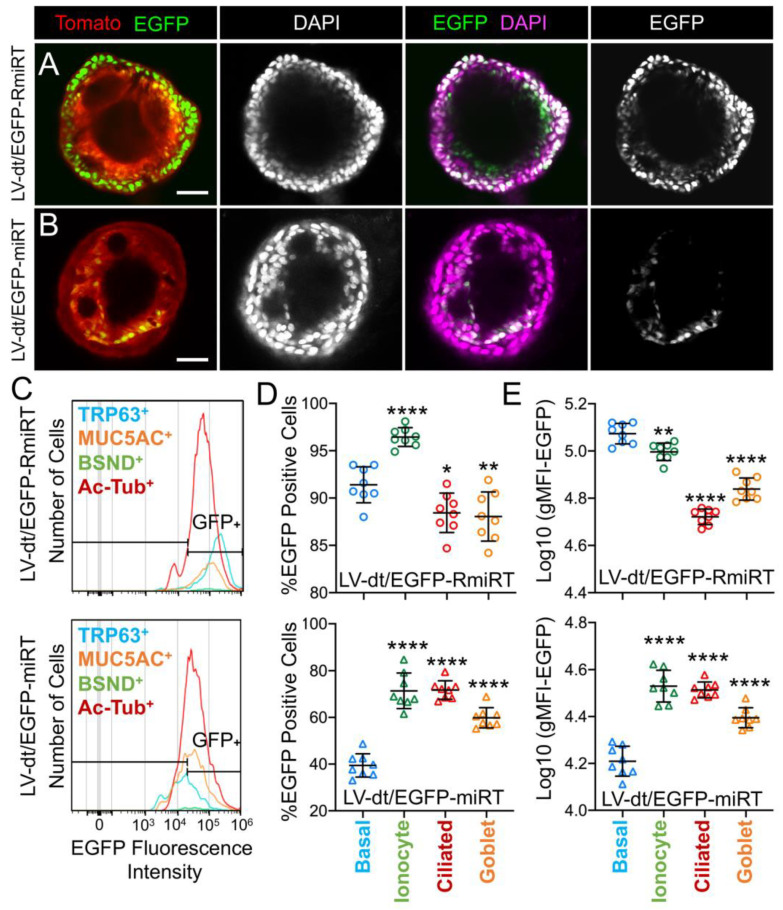
Figure 5.
Basal cell detargeting of a reporter transgene in differentiated cell types of ALI cultures. (A,B) Human basal cells were transduced with (A) LV-dt/EGFP-RmiRT or (B) LV-dt/EGFP-miRT and expanded for 1–2 days prior to seeding in organoid culture. Confocal microscopic images of live organoids stained with the Hoescht 33342 nuclei marker. Single and dual channel images are pseudocolored to better project nuclear EGFP expression. Scale bar, 50μm. (C–E) Primary basal cells were transduced with LV-dt/EGFP-RmiRT or LV-dt/EGFP-miRT viruses and then FACS was used to isolated pure Tomato-positive basal cells. These cells were expanded in culture and then seeded into ALI cultures for differentiation and then detached and immunostained for quantification of EGFP expression in various cells types by flow cytometer. (C) Epithelial lineages were stained for TRP63/p63 (basal cells; blue), BSND (ionocytes; green), alpha-tubulin (ciliated), and MUC5AC (goblet cells; orange). Representative histogram distributions of lineage-labeled cell populations treated transduced with LV-dt/EGFP-RmiRT (top) or LV-dt/EGFP-miRT (bottom). (D) Percentage of EGFP-positive cells for each lineage using the gate shown in (C) which captures 90% of EGFP-positive basal cells in the control LV-dt/EGFP-RmiRT vector group. (E) Mean fluorescent intensity (MFI) of lineage-labeled populations. Statistics represent a one-way ANOVA with Dunnett’s multiple comparison test against the basal cell population: * p < 0.01, ** p < 0.05, **** p < 0.0001. Data show the mean +/-SD for N = 8 transwells for each condition.